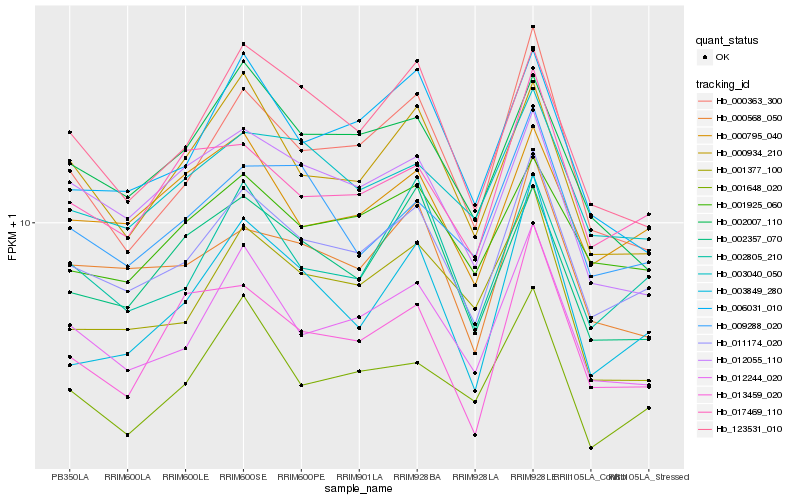
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011174_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105632610 [Jatropha curcas] |
| 2 |
Hb_012244_020 |
0.0458003606 |
- |
- |
calpain, putative [Ricinus communis] |
| 3 |
Hb_003040_050 |
0.0563025109 |
- |
- |
PREDICTED: uncharacterized protein LOC105638219 [Jatropha curcas] |
| 4 |
Hb_000568_050 |
0.0563978432 |
- |
- |
PREDICTED: endoplasmic reticulum metallopeptidase 1 isoform X2 [Jatropha curcas] |
| 5 |
Hb_002805_210 |
0.0569229956 |
transcription factor |
TF Family: SNF2 |
Chromodomain-helicase-DNA-binding 2 [Gossypium arboreum] |
| 6 |
Hb_001925_060 |
0.0575762318 |
- |
- |
PREDICTED: putative RNA polymerase II subunit B1 CTD phosphatase RPAP2 homolog [Jatropha curcas] |
| 7 |
Hb_017469_110 |
0.0591687489 |
- |
- |
PREDICTED: uncharacterized protein LOC105641795 [Jatropha curcas] |
| 8 |
Hb_000795_040 |
0.060133997 |
- |
- |
PREDICTED: sister chromatid cohesion protein PDS5 homolog A isoform X1 [Jatropha curcas] |
| 9 |
Hb_000363_300 |
0.0626564465 |
- |
- |
PREDICTED: protein ENHANCED DOWNY MILDEW 2 isoform X1 [Jatropha curcas] |
| 10 |
Hb_009288_020 |
0.0635528044 |
- |
- |
PREDICTED: protein FRIGIDA [Jatropha curcas] |
| 11 |
Hb_001648_020 |
0.0635786543 |
- |
- |
hypothetical protein POPTR_0007s07750g [Populus trichocarpa] |
| 12 |
Hb_000934_210 |
0.0640792578 |
- |
- |
suppressor of ty, putative [Ricinus communis] |
| 13 |
Hb_123531_010 |
0.0643723619 |
- |
- |
PREDICTED: nuclear pore complex protein NUP98A isoform X1 [Jatropha curcas] |
| 14 |
Hb_002357_070 |
0.0659197241 |
transcription factor |
TF Family: PHD |
PREDICTED: protein strawberry notch homolog 1 [Jatropha curcas] |
| 15 |
Hb_012055_110 |
0.0679992383 |
- |
- |
tetratricopeptide repeat protein, tpr, putative [Ricinus communis] |
| 16 |
Hb_001377_100 |
0.0695298104 |
- |
- |
PREDICTED: uncharacterized protein LOC105642863 isoform X1 [Jatropha curcas] |
| 17 |
Hb_006031_010 |
0.0697761546 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 15b isoform X3 [Jatropha curcas] |
| 18 |
Hb_002007_110 |
0.0701459871 |
transcription factor |
TF Family: Jumonji |
PREDICTED: lysine-specific demethylase JMJ25 isoform X2 [Jatropha curcas] |
| 19 |
Hb_013459_020 |
0.0704707915 |
- |
- |
PREDICTED: uncharacterized protein LOC105628886 [Jatropha curcas] |
| 20 |
Hb_003849_280 |
0.0710113814 |
- |
- |
PREDICTED: uncharacterized protein LOC105644977 [Jatropha curcas] |