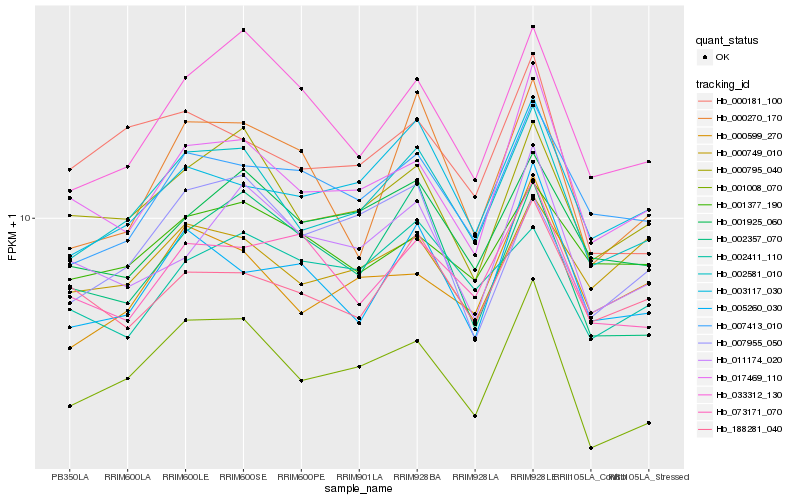
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002581_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105641469 isoform X2 [Jatropha curcas] |
| 2 |
Hb_001008_070 |
0.0462970256 |
- |
- |
PREDICTED: periodic tryptophan protein 2 homolog [Jatropha curcas] |
| 3 |
Hb_000795_040 |
0.0571986333 |
- |
- |
PREDICTED: sister chromatid cohesion protein PDS5 homolog A isoform X1 [Jatropha curcas] |
| 4 |
Hb_003117_030 |
0.0622223457 |
- |
- |
PREDICTED: protein RCC2 homolog [Jatropha curcas] |
| 5 |
Hb_017469_110 |
0.0647108085 |
- |
- |
PREDICTED: uncharacterized protein LOC105641795 [Jatropha curcas] |
| 6 |
Hb_000599_270 |
0.0653828074 |
- |
- |
PREDICTED: serine/threonine protein phosphatase 2A regulatory subunit B''beta isoform X3 [Jatropha curcas] |
| 7 |
Hb_007413_010 |
0.06659944 |
- |
- |
PREDICTED: SUMO-activating enzyme subunit 2 [Jatropha curcas] |
| 8 |
Hb_001925_060 |
0.0668281069 |
- |
- |
PREDICTED: putative RNA polymerase II subunit B1 CTD phosphatase RPAP2 homolog [Jatropha curcas] |
| 9 |
Hb_033312_130 |
0.0702312521 |
- |
- |
PREDICTED: protein TIC 40, chloroplastic [Jatropha curcas] |
| 10 |
Hb_005260_030 |
0.0749150668 |
- |
- |
PREDICTED: uncharacterized protein LOC105633782 [Jatropha curcas] |
| 11 |
Hb_001377_190 |
0.0773597689 |
transcription factor |
TF Family: PHD |
PREDICTED: histone-lysine N-methyltransferase ATX3 isoform X2 [Jatropha curcas] |
| 12 |
Hb_002357_070 |
0.0773722959 |
transcription factor |
TF Family: PHD |
PREDICTED: protein strawberry notch homolog 1 [Jatropha curcas] |
| 13 |
Hb_188281_040 |
0.0777956791 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 12 [Jatropha curcas] |
| 14 |
Hb_011174_020 |
0.0792699518 |
- |
- |
PREDICTED: uncharacterized protein LOC105632610 [Jatropha curcas] |
| 15 |
Hb_000749_010 |
0.0796778662 |
- |
- |
PREDICTED: non-canonical poly(A) RNA polymerase PAPD5 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000181_100 |
0.0797181563 |
- |
- |
PREDICTED: protein starmaker [Jatropha curcas] |
| 17 |
Hb_007955_050 |
0.0805952085 |
- |
- |
PREDICTED: multiple RNA-binding domain-containing protein 1 [Jatropha curcas] |
| 18 |
Hb_002411_110 |
0.0810975594 |
- |
- |
PREDICTED: DNA-directed RNA polymerase III subunit RPC2 [Jatropha curcas] |
| 19 |
Hb_073171_070 |
0.0819667698 |
- |
- |
PREDICTED: RNA polymerase II-associated factor 1 homolog [Jatropha curcas] |
| 20 |
Hb_000270_170 |
0.0824520906 |
- |
- |
PREDICTED: formin-like protein 5 [Jatropha curcas] |