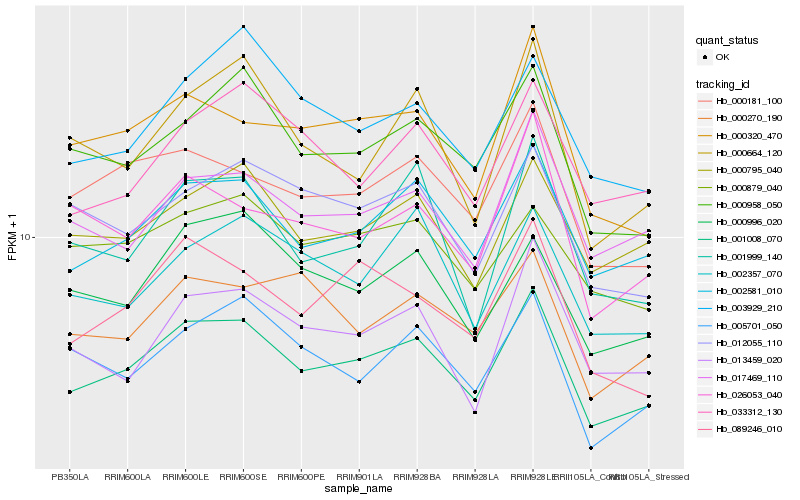
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001008_070 |
0.0 |
- |
- |
PREDICTED: periodic tryptophan protein 2 homolog [Jatropha curcas] |
| 2 |
Hb_002581_010 |
0.0462970256 |
- |
- |
PREDICTED: uncharacterized protein LOC105641469 isoform X2 [Jatropha curcas] |
| 3 |
Hb_000181_100 |
0.0653908731 |
- |
- |
PREDICTED: protein starmaker [Jatropha curcas] |
| 4 |
Hb_017469_110 |
0.0663527403 |
- |
- |
PREDICTED: uncharacterized protein LOC105641795 [Jatropha curcas] |
| 5 |
Hb_000795_040 |
0.0664222352 |
- |
- |
PREDICTED: sister chromatid cohesion protein PDS5 homolog A isoform X1 [Jatropha curcas] |
| 6 |
Hb_000958_050 |
0.0665453747 |
- |
- |
hypothetical protein JCGZ_16781 [Jatropha curcas] |
| 7 |
Hb_000320_470 |
0.0710543344 |
- |
- |
calcium-dependent protein kinase [Hevea brasiliensis] |
| 8 |
Hb_089246_010 |
0.0736282768 |
- |
- |
hypothetical protein JCGZ_24395 [Jatropha curcas] |
| 9 |
Hb_000996_020 |
0.0747494784 |
- |
- |
PREDICTED: RNA-binding protein NOB1 [Jatropha curcas] |
| 10 |
Hb_012055_110 |
0.0750319477 |
- |
- |
tetratricopeptide repeat protein, tpr, putative [Ricinus communis] |
| 11 |
Hb_013459_020 |
0.0753091456 |
- |
- |
PREDICTED: uncharacterized protein LOC105628886 [Jatropha curcas] |
| 12 |
Hb_005701_050 |
0.0774253679 |
- |
- |
PREDICTED: protein LTV1 homolog [Jatropha curcas] |
| 13 |
Hb_033312_130 |
0.07744195 |
- |
- |
PREDICTED: protein TIC 40, chloroplastic [Jatropha curcas] |
| 14 |
Hb_002357_070 |
0.0783877022 |
transcription factor |
TF Family: PHD |
PREDICTED: protein strawberry notch homolog 1 [Jatropha curcas] |
| 15 |
Hb_000879_040 |
0.0795059287 |
- |
- |
PREDICTED: calcium-transporting ATPase 3, endoplasmic reticulum-type [Jatropha curcas] |
| 16 |
Hb_003929_210 |
0.079852096 |
transcription factor |
TF Family: SBP |
Squamosa promoter-binding protein, putative [Ricinus communis] |
| 17 |
Hb_026053_040 |
0.0800141218 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000664_120 |
0.0803106694 |
- |
- |
PREDICTED: SART-1 family protein DOT2 isoform X1 [Jatropha curcas] |
| 19 |
Hb_001999_140 |
0.0805368843 |
- |
- |
PREDICTED: anthranilate synthase alpha subunit 2, chloroplastic isoform X2 [Jatropha curcas] |
| 20 |
Hb_000270_190 |
0.0817684399 |
transcription factor |
TF Family: DDT |
tRNA ligase, putative [Ricinus communis] |