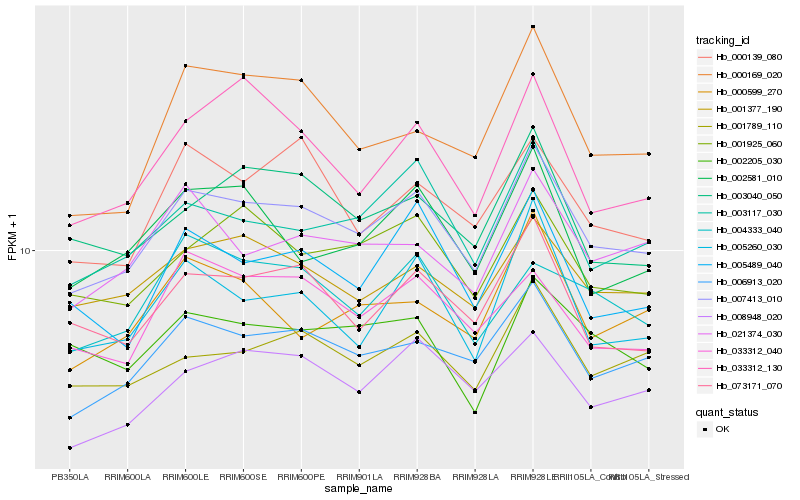
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007413_010 |
0.0 |
- |
- |
PREDICTED: SUMO-activating enzyme subunit 2 [Jatropha curcas] |
| 2 |
Hb_006913_020 |
0.0444640334 |
- |
- |
PREDICTED: uncharacterized protein LOC105649145 isoform X1 [Jatropha curcas] |
| 3 |
Hb_003117_030 |
0.0544555437 |
- |
- |
PREDICTED: protein RCC2 homolog [Jatropha curcas] |
| 4 |
Hb_008948_020 |
0.0566117739 |
- |
- |
hypothetical protein JCGZ_21216 [Jatropha curcas] |
| 5 |
Hb_001377_190 |
0.0572953796 |
transcription factor |
TF Family: PHD |
PREDICTED: histone-lysine N-methyltransferase ATX3 isoform X2 [Jatropha curcas] |
| 6 |
Hb_001925_060 |
0.0575347645 |
- |
- |
PREDICTED: putative RNA polymerase II subunit B1 CTD phosphatase RPAP2 homolog [Jatropha curcas] |
| 7 |
Hb_001789_110 |
0.0575443053 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 8 |
Hb_073171_070 |
0.0583778285 |
- |
- |
PREDICTED: RNA polymerase II-associated factor 1 homolog [Jatropha curcas] |
| 9 |
Hb_005260_030 |
0.0614385759 |
- |
- |
PREDICTED: uncharacterized protein LOC105633782 [Jatropha curcas] |
| 10 |
Hb_005489_040 |
0.062944034 |
- |
- |
PREDICTED: cullin-1 [Jatropha curcas] |
| 11 |
Hb_021374_030 |
0.0639949742 |
- |
- |
hypothetical protein RCOM_0351490 [Ricinus communis] |
| 12 |
Hb_003040_050 |
0.0655956201 |
- |
- |
PREDICTED: uncharacterized protein LOC105638219 [Jatropha curcas] |
| 13 |
Hb_000139_080 |
0.0659820375 |
- |
- |
PREDICTED: vacuole membrane protein KMS1 isoform X2 [Jatropha curcas] |
| 14 |
Hb_002581_010 |
0.06659944 |
- |
- |
PREDICTED: uncharacterized protein LOC105641469 isoform X2 [Jatropha curcas] |
| 15 |
Hb_033312_040 |
0.0666912678 |
- |
- |
PREDICTED: gamma-tubulin complex component 5-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_000169_020 |
0.06817343 |
transcription factor |
TF Family: C2C2-CO-like |
hypothetical protein RCOM_0555710 [Ricinus communis] |
| 17 |
Hb_000599_270 |
0.0682686686 |
- |
- |
PREDICTED: serine/threonine protein phosphatase 2A regulatory subunit B''beta isoform X3 [Jatropha curcas] |
| 18 |
Hb_004333_040 |
0.0686889228 |
- |
- |
PREDICTED: uncharacterized protein LOC105629988 [Jatropha curcas] |
| 19 |
Hb_033312_130 |
0.0691337038 |
- |
- |
PREDICTED: protein TIC 40, chloroplastic [Jatropha curcas] |
| 20 |
Hb_002205_030 |
0.0710786828 |
- |
- |
PREDICTED: alpha-galactosidase 3 [Jatropha curcas] |