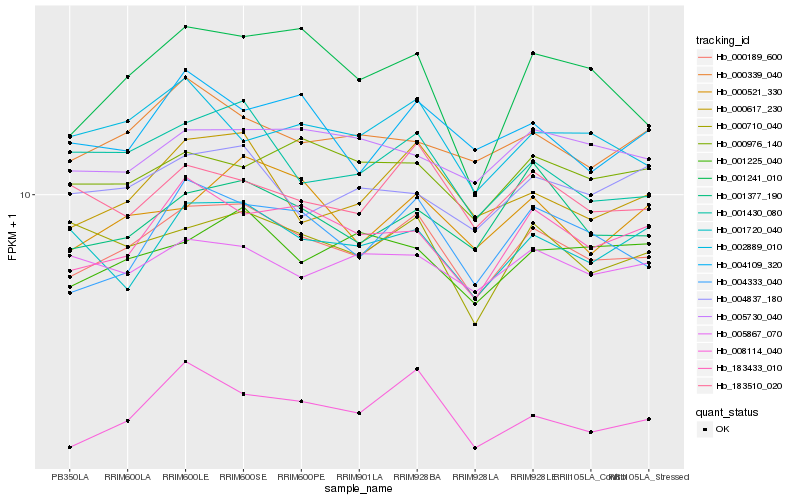
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000189_600 |
0.0 |
- |
- |
PREDICTED: protein MON2 homolog isoform X1 [Jatropha curcas] |
| 2 |
Hb_004333_040 |
0.0573092969 |
- |
- |
PREDICTED: uncharacterized protein LOC105629988 [Jatropha curcas] |
| 3 |
Hb_183433_010 |
0.0582069214 |
- |
- |
PREDICTED: FAD synthase isoform X3 [Jatropha curcas] |
| 4 |
Hb_000617_230 |
0.0589167754 |
- |
- |
PREDICTED: tafazzin isoform X1 [Jatropha curcas] |
| 5 |
Hb_001377_190 |
0.0591500015 |
transcription factor |
TF Family: PHD |
PREDICTED: histone-lysine N-methyltransferase ATX3 isoform X2 [Jatropha curcas] |
| 6 |
Hb_001430_080 |
0.0624626376 |
- |
- |
PREDICTED: putative mediator of RNA polymerase II transcription subunit 26 [Jatropha curcas] |
| 7 |
Hb_000521_330 |
0.0634403672 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_183510_020 |
0.0642234543 |
- |
- |
coated vesicle membrane protein, putative [Ricinus communis] |
| 9 |
Hb_000710_040 |
0.0651301316 |
- |
- |
PREDICTED: HEAT repeat-containing protein 6 [Jatropha curcas] |
| 10 |
Hb_001225_040 |
0.0657993005 |
- |
- |
PREDICTED: meiosis-specific nuclear structural protein 1 isoform X5 [Jatropha curcas] |
| 11 |
Hb_000339_040 |
0.0667392995 |
- |
- |
PREDICTED: SAP30-binding protein isoform X2 [Jatropha curcas] |
| 12 |
Hb_005730_040 |
0.06728633 |
- |
- |
hypothetical protein PRUPE_ppa004338mg [Prunus persica] |
| 13 |
Hb_000976_140 |
0.0673623593 |
- |
- |
PREDICTED: uncharacterized protein LOC105646208 isoform X1 [Jatropha curcas] |
| 14 |
Hb_008114_040 |
0.0679026004 |
- |
- |
Fanconi anemia group D2 [Gossypium arboreum] |
| 15 |
Hb_002889_010 |
0.0683731704 |
- |
- |
ubx domain-containing, putative [Ricinus communis] |
| 16 |
Hb_005867_070 |
0.0689565261 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 17 |
Hb_004109_320 |
0.0694995192 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 52 A [Jatropha curcas] |
| 18 |
Hb_001241_010 |
0.070376912 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 19 |
Hb_004837_180 |
0.0704331999 |
transcription factor |
TF Family: NF-YA |
PREDICTED: nuclear transcription factor Y subunit A-1-like isoform X1 [Jatropha curcas] |
| 20 |
Hb_001720_040 |
0.0710662574 |
- |
- |
conserved hypothetical protein [Ricinus communis] |