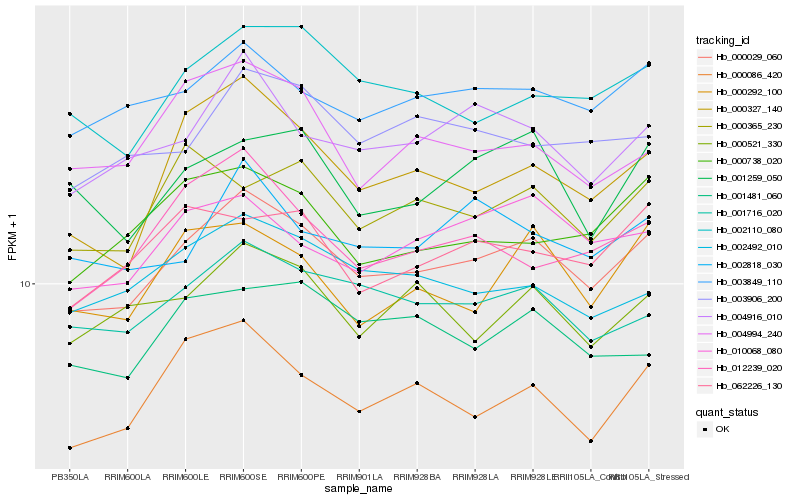
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000029_060 |
0.0 |
- |
- |
PREDICTED: mRNA-decapping enzyme-like protein [Jatropha curcas] |
| 2 |
Hb_003849_110 |
0.0554126927 |
- |
- |
hypothetical protein JCGZ_17842 [Jatropha curcas] |
| 3 |
Hb_001716_020 |
0.055667779 |
- |
- |
hydrolase, putative [Ricinus communis] |
| 4 |
Hb_004916_010 |
0.058565077 |
- |
- |
PREDICTED: uncharacterized protein LOC105631086 [Jatropha curcas] |
| 5 |
Hb_002110_080 |
0.0601406073 |
- |
- |
FH interacting protein 1 [Theobroma cacao] |
| 6 |
Hb_000086_420 |
0.061827309 |
- |
- |
PREDICTED: phospholipid--sterol O-acyltransferase [Jatropha curcas] |
| 7 |
Hb_000327_140 |
0.0620840319 |
- |
- |
PREDICTED: probable adenylate kinase 6, chloroplastic [Jatropha curcas] |
| 8 |
Hb_002492_010 |
0.064717216 |
- |
- |
PREDICTED: inositol-tetrakisphosphate 1-kinase 2-like [Populus euphratica] |
| 9 |
Hb_010068_080 |
0.0649341016 |
- |
- |
PREDICTED: RNA-binding protein rsd1-like [Jatropha curcas] |
| 10 |
Hb_003906_200 |
0.0657412097 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 48 [Jatropha curcas] |
| 11 |
Hb_000521_330 |
0.0658335354 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000738_020 |
0.0669422015 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 13 |
Hb_012239_020 |
0.0673276925 |
- |
- |
PREDICTED: protein VAC14 homolog [Jatropha curcas] |
| 14 |
Hb_062226_130 |
0.0679157438 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase PP1 isozyme 4 [Jatropha curcas] |
| 15 |
Hb_000292_100 |
0.0680224481 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor HBP-1a [Jatropha curcas] |
| 16 |
Hb_002818_030 |
0.0681160544 |
- |
- |
PREDICTED: protein transport protein SEC23 [Jatropha curcas] |
| 17 |
Hb_001259_050 |
0.0691166706 |
- |
- |
calcium lipid binding protein, putative [Ricinus communis] |
| 18 |
Hb_001481_060 |
0.0707135173 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase HOS1 isoform X1 [Jatropha curcas] |
| 19 |
Hb_004994_240 |
0.0712045852 |
- |
- |
hypothetical protein PRUPE_ppa004633mg [Prunus persica] |
| 20 |
Hb_000365_230 |
0.0726388066 |
- |
- |
PREDICTED: uncharacterized protein LOC105649056 [Jatropha curcas] |