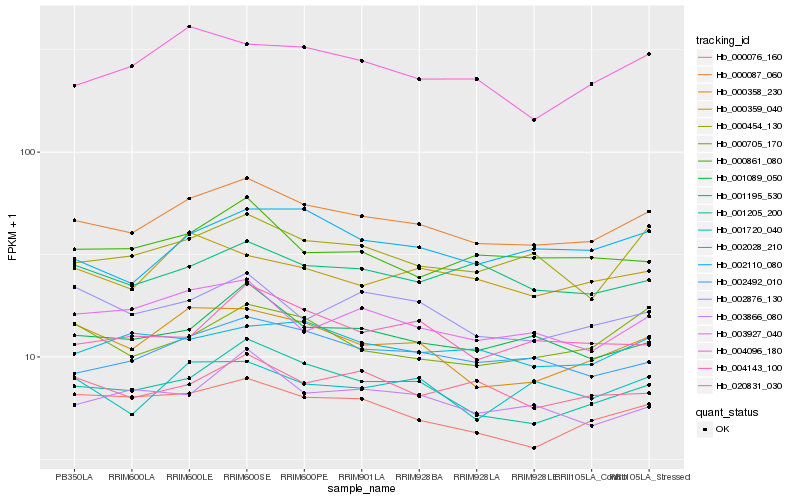
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000087_060 |
0.0 |
- |
- |
protein transporter, putative [Ricinus communis] |
| 2 |
Hb_001205_200 |
0.0462428677 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_002492_010 |
0.05484306 |
- |
- |
PREDICTED: inositol-tetrakisphosphate 1-kinase 2-like [Populus euphratica] |
| 4 |
Hb_001089_050 |
0.0572533148 |
transcription factor |
TF Family: ARID |
PREDICTED: AT-rich interactive domain-containing protein 4 [Jatropha curcas] |
| 5 |
Hb_000358_230 |
0.0577885045 |
- |
- |
protein phosphatases pp1 regulatory subunit, putative [Ricinus communis] |
| 6 |
Hb_000076_160 |
0.0603427433 |
- |
- |
PREDICTED: AP-5 complex subunit zeta-1 [Jatropha curcas] |
| 7 |
Hb_000454_130 |
0.0605052135 |
transcription factor |
TF Family: BBR-BPC |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_003927_040 |
0.0618581843 |
- |
- |
CCR4-NOT transcription complex subunit, putative [Ricinus communis] |
| 9 |
Hb_001195_530 |
0.064291906 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_002110_080 |
0.0656485226 |
- |
- |
FH interacting protein 1 [Theobroma cacao] |
| 11 |
Hb_003866_080 |
0.0671738238 |
- |
- |
PREDICTED: uncharacterized protein LOC105639150 [Jatropha curcas] |
| 12 |
Hb_002028_210 |
0.0675564027 |
- |
- |
PREDICTED: uncharacterized protein LOC105638038 [Jatropha curcas] |
| 13 |
Hb_004096_180 |
0.0676338918 |
- |
- |
PREDICTED: 14-3-3-like protein A [Jatropha curcas] |
| 14 |
Hb_020831_030 |
0.0678527801 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_002876_130 |
0.0683692833 |
- |
- |
PREDICTED: uncharacterized protein LOC105637143 [Jatropha curcas] |
| 16 |
Hb_001720_040 |
0.0686031111 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000705_170 |
0.0692649448 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g46870 [Jatropha curcas] |
| 18 |
Hb_000359_040 |
0.069678566 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_004143_100 |
0.0697663177 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000861_080 |
0.0706476021 |
- |
- |
PREDICTED: protein ELC-like [Jatropha curcas] |