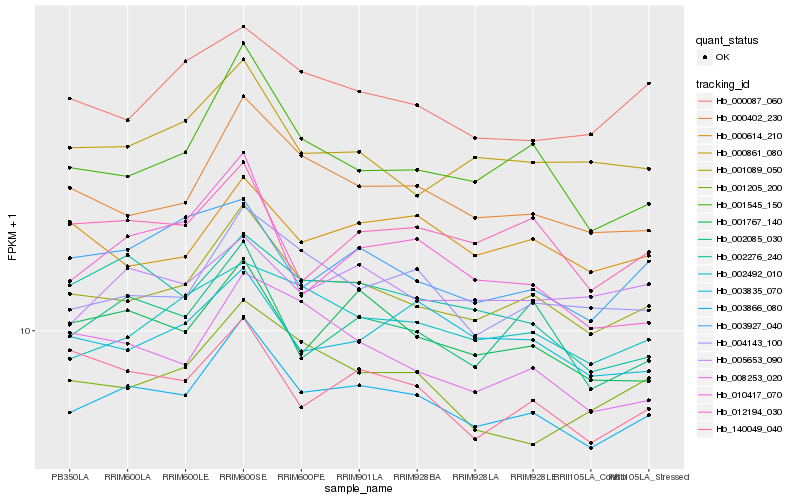
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003866_080 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105639150 [Jatropha curcas] |
| 2 |
Hb_001089_050 |
0.0336894892 |
transcription factor |
TF Family: ARID |
PREDICTED: AT-rich interactive domain-containing protein 4 [Jatropha curcas] |
| 3 |
Hb_000402_230 |
0.0530721459 |
- |
- |
PREDICTED: sorting nexin 2A-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_004143_100 |
0.0535969659 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_001767_140 |
0.055278044 |
- |
- |
PREDICTED: protein-tyrosine-phosphatase MKP1 [Jatropha curcas] |
| 6 |
Hb_002085_030 |
0.0580995289 |
- |
- |
PREDICTED: protein CHROMATIN REMODELING 4 isoform X2 [Jatropha curcas] |
| 7 |
Hb_002492_010 |
0.0602811062 |
- |
- |
PREDICTED: inositol-tetrakisphosphate 1-kinase 2-like [Populus euphratica] |
| 8 |
Hb_001545_150 |
0.0616759474 |
transcription factor |
TF Family: NAC |
PREDICTED: uncharacterized protein LOC105635527 [Jatropha curcas] |
| 9 |
Hb_002276_240 |
0.0629205206 |
- |
- |
PREDICTED: uncharacterized protein LOC105640682 [Jatropha curcas] |
| 10 |
Hb_003835_070 |
0.0637357842 |
- |
- |
PREDICTED: uncharacterized protein LOC105641698 isoform X2 [Jatropha curcas] |
| 11 |
Hb_140049_040 |
0.0637516789 |
- |
- |
PREDICTED: uncharacterized protein LOC105632012 [Jatropha curcas] |
| 12 |
Hb_012194_030 |
0.0652389097 |
- |
- |
PREDICTED: R3H domain-containing protein 2-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_003927_040 |
0.0657189072 |
- |
- |
CCR4-NOT transcription complex subunit, putative [Ricinus communis] |
| 14 |
Hb_000861_080 |
0.0664357204 |
- |
- |
PREDICTED: protein ELC-like [Jatropha curcas] |
| 15 |
Hb_000614_210 |
0.0668834992 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor GT-1-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_005653_090 |
0.0670311319 |
- |
- |
PREDICTED: protein HIRA isoform X1 [Jatropha curcas] |
| 17 |
Hb_000087_060 |
0.0671738238 |
- |
- |
protein transporter, putative [Ricinus communis] |
| 18 |
Hb_001205_200 |
0.0683707439 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_008253_020 |
0.0683772558 |
- |
- |
PREDICTED: serine/threonine protein phosphatase 2A 57 kDa regulatory subunit B' theta isoform-like [Jatropha curcas] |
| 20 |
Hb_010417_070 |
0.0685918364 |
- |
- |
PREDICTED: uncharacterized protein LOC105645744 isoform X1 [Jatropha curcas] |