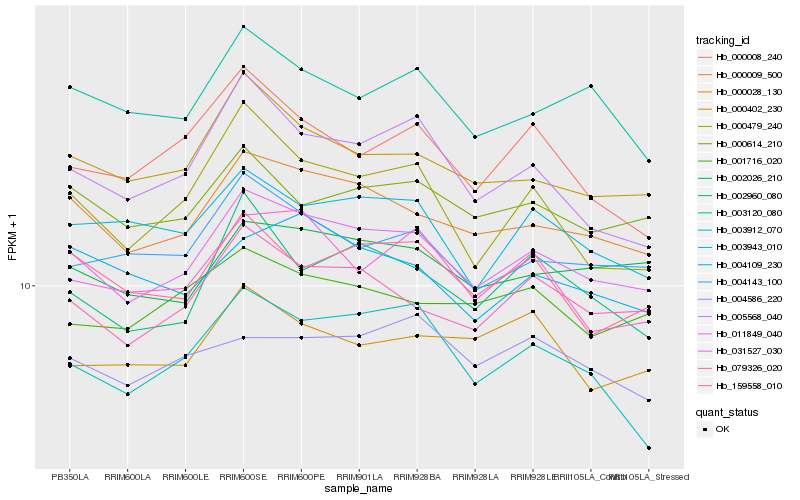
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011849_040 |
0.0 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 2 |
Hb_000009_500 |
0.034974383 |
- |
- |
PREDICTED: NASP-related protein sim3 [Jatropha curcas] |
| 3 |
Hb_079326_020 |
0.0476594586 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000402_230 |
0.053190551 |
- |
- |
PREDICTED: sorting nexin 2A-like isoform X1 [Jatropha curcas] |
| 5 |
Hb_002026_210 |
0.0578194643 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At5g41260 [Jatropha curcas] |
| 6 |
Hb_004143_100 |
0.0597665331 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000614_210 |
0.0614238086 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor GT-1-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_003120_080 |
0.0635545344 |
- |
- |
PREDICTED: COPII coat assembly protein SEC16 isoform X1 [Jatropha curcas] |
| 9 |
Hb_159558_010 |
0.0636290393 |
- |
- |
PREDICTED: uncharacterized protein LOC105635846 [Jatropha curcas] |
| 10 |
Hb_003943_010 |
0.064188864 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_002960_080 |
0.0644416366 |
transcription factor |
TF Family: IWS1 |
PREDICTED: uncharacterized protein LOC105635322 [Jatropha curcas] |
| 12 |
Hb_031527_030 |
0.0649070878 |
- |
- |
PREDICTED: ribosomal RNA processing protein 1 homolog [Jatropha curcas] |
| 13 |
Hb_005568_040 |
0.0666597357 |
- |
- |
PREDICTED: uncharacterized protein LOC105628163 isoform X2 [Jatropha curcas] |
| 14 |
Hb_001716_020 |
0.0669521332 |
- |
- |
hydrolase, putative [Ricinus communis] |
| 15 |
Hb_000479_240 |
0.069138256 |
- |
- |
PREDICTED: probable ubiquitin conjugation factor E4 isoform X2 [Jatropha curcas] |
| 16 |
Hb_004109_230 |
0.0692436567 |
- |
- |
heat shock protein binding protein, putative [Ricinus communis] |
| 17 |
Hb_003912_070 |
0.0732614323 |
- |
- |
PREDICTED: nuclear pore complex protein NUP85 [Jatropha curcas] |
| 18 |
Hb_004586_220 |
0.0735511197 |
- |
- |
PREDICTED: WD and tetratricopeptide repeats protein 1 isoform X3 [Jatropha curcas] |
| 19 |
Hb_000008_240 |
0.0741897944 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000028_130 |
0.0746546181 |
- |
- |
PREDICTED: uncharacterized protein LOC105645303 isoform X1 [Jatropha curcas] |