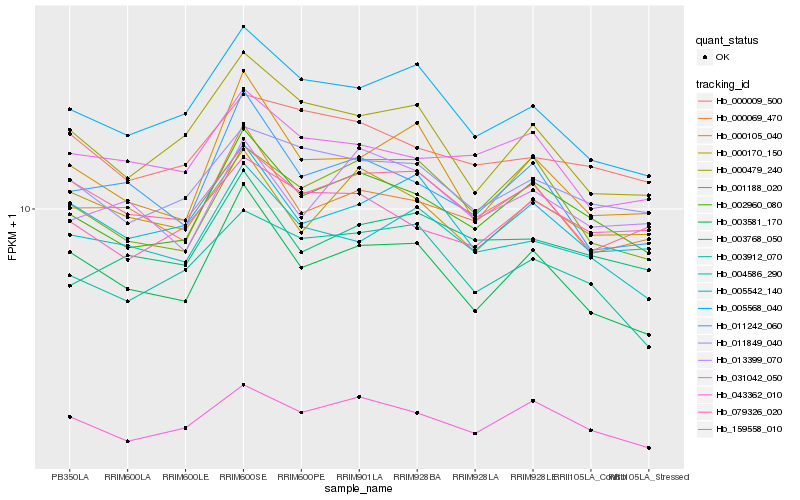
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002960_080 |
0.0 |
transcription factor |
TF Family: IWS1 |
PREDICTED: uncharacterized protein LOC105635322 [Jatropha curcas] |
| 2 |
Hb_043362_010 |
0.0543693125 |
- |
- |
hypothetical protein JCGZ_23765 [Jatropha curcas] |
| 3 |
Hb_079326_020 |
0.0639264017 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_011849_040 |
0.0644416366 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 5 |
Hb_000069_470 |
0.0689077269 |
- |
- |
PREDICTED: uncharacterized protein LOC105642863 isoform X1 [Jatropha curcas] |
| 6 |
Hb_001188_020 |
0.070847499 |
- |
- |
PREDICTED: E3 SUMO-protein ligase SIZ1 isoform X1 [Jatropha curcas] |
| 7 |
Hb_003581_170 |
0.0722563283 |
- |
- |
PREDICTED: alpha-amylase 3, chloroplastic [Jatropha curcas] |
| 8 |
Hb_005542_140 |
0.0767415998 |
- |
- |
PREDICTED: U-box domain-containing protein 62-like [Jatropha curcas] |
| 9 |
Hb_004586_290 |
0.0779033479 |
- |
- |
PREDICTED: transport inhibitor response 1-like protein [Jatropha curcas] |
| 10 |
Hb_003768_050 |
0.0779200845 |
- |
- |
PREDICTED: uncharacterized protein LOC105644152 isoform X1 [Jatropha curcas] |
| 11 |
Hb_031042_050 |
0.0780643305 |
- |
- |
PREDICTED: protein transport protein SEC16B homolog [Jatropha curcas] |
| 12 |
Hb_005568_040 |
0.0783068926 |
- |
- |
PREDICTED: uncharacterized protein LOC105628163 isoform X2 [Jatropha curcas] |
| 13 |
Hb_000009_500 |
0.078478368 |
- |
- |
PREDICTED: NASP-related protein sim3 [Jatropha curcas] |
| 14 |
Hb_159558_010 |
0.0791892422 |
- |
- |
PREDICTED: uncharacterized protein LOC105635846 [Jatropha curcas] |
| 15 |
Hb_000170_150 |
0.0795595615 |
- |
- |
PREDICTED: uncharacterized protein LOC105638996 isoform X3 [Jatropha curcas] |
| 16 |
Hb_011242_060 |
0.0796354288 |
transcription factor |
TF Family: SNF2 |
Chromo domain protein, putative [Ricinus communis] |
| 17 |
Hb_003912_070 |
0.0807300627 |
- |
- |
PREDICTED: nuclear pore complex protein NUP85 [Jatropha curcas] |
| 18 |
Hb_013399_070 |
0.0807827331 |
- |
- |
PREDICTED: uncharacterized protein LOC105631409 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000105_040 |
0.0817246847 |
- |
- |
PREDICTED: serine/threonine-protein kinase TOR isoform X2 [Jatropha curcas] |
| 20 |
Hb_000479_240 |
0.0825709043 |
- |
- |
PREDICTED: probable ubiquitin conjugation factor E4 isoform X2 [Jatropha curcas] |