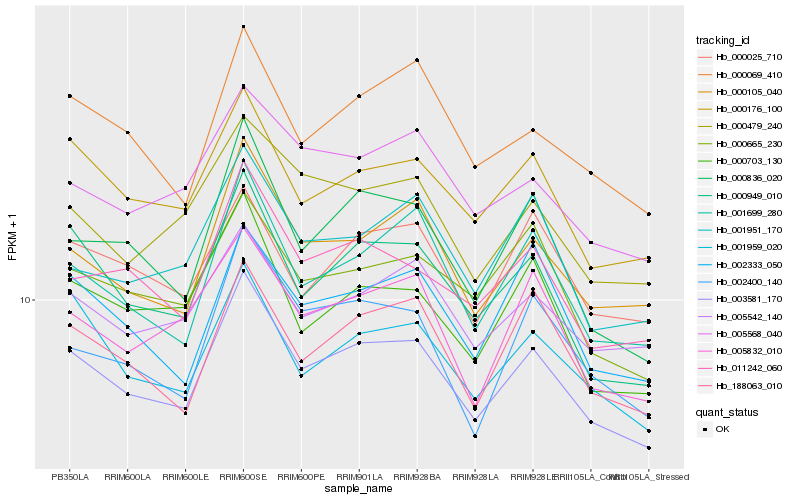
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003581_170 |
0.0 |
- |
- |
PREDICTED: alpha-amylase 3, chloroplastic [Jatropha curcas] |
| 2 |
Hb_000105_040 |
0.0523856452 |
- |
- |
PREDICTED: serine/threonine-protein kinase TOR isoform X2 [Jatropha curcas] |
| 3 |
Hb_000176_100 |
0.054004024 |
- |
- |
PREDICTED: probable splicing factor 3A subunit 1 [Solanum lycopersicum] |
| 4 |
Hb_000949_010 |
0.0554617262 |
- |
- |
PREDICTED: regulator of nonsense transcripts UPF2 [Jatropha curcas] |
| 5 |
Hb_001699_280 |
0.0575966012 |
- |
- |
PREDICTED: transformation/transcription domain-associated protein [Jatropha curcas] |
| 6 |
Hb_011242_060 |
0.0600412252 |
transcription factor |
TF Family: SNF2 |
Chromo domain protein, putative [Ricinus communis] |
| 7 |
Hb_000836_020 |
0.0612076686 |
transcription factor |
TF Family: SNF2 |
Chromo domain protein, putative [Ricinus communis] |
| 8 |
Hb_188063_010 |
0.0615597217 |
- |
- |
hypothetical protein RCOM_1469330 [Ricinus communis] |
| 9 |
Hb_001959_020 |
0.0617592752 |
- |
- |
PREDICTED: uncharacterized protein LOC105638454 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000069_410 |
0.0633446693 |
- |
- |
PREDICTED: protein SDE2 homolog [Jatropha curcas] |
| 11 |
Hb_005542_140 |
0.0656034719 |
- |
- |
PREDICTED: U-box domain-containing protein 62-like [Jatropha curcas] |
| 12 |
Hb_000665_230 |
0.0660062178 |
- |
- |
PREDICTED: uncharacterized protein LOC105637615 [Jatropha curcas] |
| 13 |
Hb_005832_010 |
0.0660471979 |
- |
- |
PREDICTED: enhancer of mRNA-decapping protein 4 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000703_130 |
0.0664875115 |
- |
- |
PREDICTED: ATP-dependent RNA helicase DHX36 [Jatropha curcas] |
| 15 |
Hb_005568_040 |
0.0667526962 |
- |
- |
PREDICTED: uncharacterized protein LOC105628163 isoform X2 [Jatropha curcas] |
| 16 |
Hb_000479_240 |
0.0674263677 |
- |
- |
PREDICTED: probable ubiquitin conjugation factor E4 isoform X2 [Jatropha curcas] |
| 17 |
Hb_002333_050 |
0.0691210949 |
- |
- |
PREDICTED: uncharacterized protein LOC105644854 isoform X1 [Jatropha curcas] |
| 18 |
Hb_002400_140 |
0.0692131217 |
- |
- |
protein kinase, putative [Ricinus communis] |
| 19 |
Hb_000025_710 |
0.0699872024 |
transcription factor |
TF Family: SNF2 |
Chromodomain-helicase-DNA-binding 2 [Gossypium arboreum] |
| 20 |
Hb_001951_170 |
0.0702521904 |
- |
- |
PREDICTED: uncharacterized protein LOC105649755 [Jatropha curcas] |