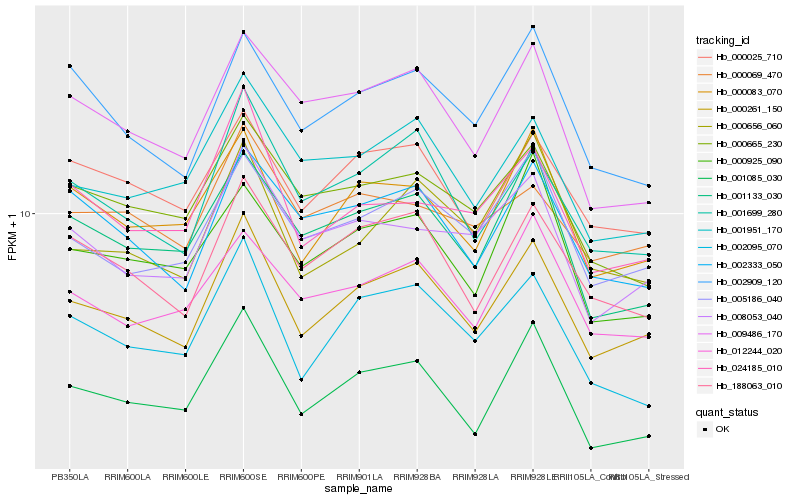
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000261_150 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_001699_280 |
0.0618531515 |
- |
- |
PREDICTED: transformation/transcription domain-associated protein [Jatropha curcas] |
| 3 |
Hb_188063_010 |
0.062409523 |
- |
- |
hypothetical protein RCOM_1469330 [Ricinus communis] |
| 4 |
Hb_000083_070 |
0.0644507258 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 1 isoform X2 [Jatropha curcas] |
| 5 |
Hb_005186_040 |
0.0644825849 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 13 isoform X2 [Jatropha curcas] |
| 6 |
Hb_000025_710 |
0.0699757882 |
transcription factor |
TF Family: SNF2 |
Chromodomain-helicase-DNA-binding 2 [Gossypium arboreum] |
| 7 |
Hb_002095_070 |
0.0726735433 |
- |
- |
PREDICTED: probable pre-mRNA-splicing factor ATP-dependent RNA helicase [Jatropha curcas] |
| 8 |
Hb_002333_050 |
0.0730079534 |
- |
- |
PREDICTED: uncharacterized protein LOC105644854 isoform X1 [Jatropha curcas] |
| 9 |
Hb_001133_030 |
0.0730170405 |
- |
- |
PREDICTED: MAG2-interacting protein 2-like [Jatropha curcas] |
| 10 |
Hb_001951_170 |
0.074427558 |
- |
- |
PREDICTED: uncharacterized protein LOC105649755 [Jatropha curcas] |
| 11 |
Hb_002909_120 |
0.0747603717 |
- |
- |
PREDICTED: transcription elongation factor SPT6 [Jatropha curcas] |
| 12 |
Hb_012244_020 |
0.0753814282 |
- |
- |
calpain, putative [Ricinus communis] |
| 13 |
Hb_008053_040 |
0.0755663144 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 12-like [Pyrus x bretschneideri] |
| 14 |
Hb_009486_170 |
0.0762372764 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 4G [Jatropha curcas] |
| 15 |
Hb_000656_060 |
0.0763158489 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 16 |
Hb_001085_030 |
0.0775950218 |
- |
- |
Tetratricopeptide repeat-like superfamily protein, putative isoform 1 [Theobroma cacao] |
| 17 |
Hb_024185_010 |
0.0794880738 |
transcription factor |
TF Family: PHD |
PREDICTED: histone acetyltransferase HAC1 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000665_230 |
0.0827673009 |
- |
- |
PREDICTED: uncharacterized protein LOC105637615 [Jatropha curcas] |
| 19 |
Hb_000925_090 |
0.0830764724 |
- |
- |
PREDICTED: uncharacterized protein LOC105635805 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000069_470 |
0.085788455 |
- |
- |
PREDICTED: uncharacterized protein LOC105642863 isoform X1 [Jatropha curcas] |