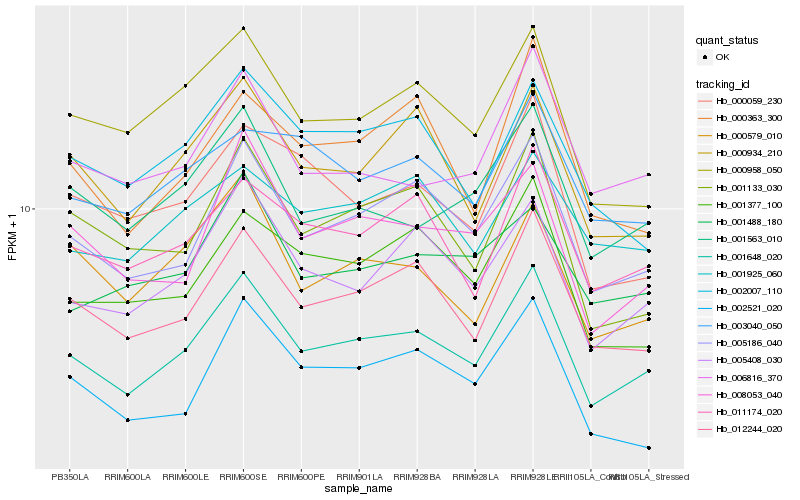
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001648_020 |
0.0 |
- |
- |
hypothetical protein POPTR_0007s07750g [Populus trichocarpa] |
| 2 |
Hb_005186_040 |
0.0484504278 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 13 isoform X2 [Jatropha curcas] |
| 3 |
Hb_012244_020 |
0.0510962182 |
- |
- |
calpain, putative [Ricinus communis] |
| 4 |
Hb_005408_030 |
0.0577415251 |
- |
- |
PREDICTED: uncharacterized protein LOC105648043 [Jatropha curcas] |
| 5 |
Hb_011174_020 |
0.0635786543 |
- |
- |
PREDICTED: uncharacterized protein LOC105632610 [Jatropha curcas] |
| 6 |
Hb_000934_210 |
0.069291678 |
- |
- |
suppressor of ty, putative [Ricinus communis] |
| 7 |
Hb_001563_010 |
0.0698078819 |
transcription factor |
TF Family: SET |
PREDICTED: probable histone-lysine N-methyltransferase ATXR3 [Jatropha curcas] |
| 8 |
Hb_000958_050 |
0.0703394749 |
- |
- |
hypothetical protein JCGZ_16781 [Jatropha curcas] |
| 9 |
Hb_008053_040 |
0.0709473948 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 12-like [Pyrus x bretschneideri] |
| 10 |
Hb_006816_370 |
0.0715136437 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105647456 [Jatropha curcas] |
| 11 |
Hb_001377_100 |
0.0746802912 |
- |
- |
PREDICTED: uncharacterized protein LOC105642863 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000579_010 |
0.0748781245 |
transcription factor |
TF Family: SET |
PREDICTED: probable histone-lysine N-methyltransferase ATXR3 [Jatropha curcas] |
| 13 |
Hb_001133_030 |
0.0759496855 |
- |
- |
PREDICTED: MAG2-interacting protein 2-like [Jatropha curcas] |
| 14 |
Hb_000059_230 |
0.077997514 |
- |
- |
PREDICTED: uncharacterized protein LOC105648049 [Jatropha curcas] |
| 15 |
Hb_001488_180 |
0.0780528323 |
- |
- |
PREDICTED: uncharacterized protein LOC105644152 isoform X1 [Jatropha curcas] |
| 16 |
Hb_003040_050 |
0.0785421507 |
- |
- |
PREDICTED: uncharacterized protein LOC105638219 [Jatropha curcas] |
| 17 |
Hb_001925_060 |
0.0788503004 |
- |
- |
PREDICTED: putative RNA polymerase II subunit B1 CTD phosphatase RPAP2 homolog [Jatropha curcas] |
| 18 |
Hb_002007_110 |
0.0788619796 |
transcription factor |
TF Family: Jumonji |
PREDICTED: lysine-specific demethylase JMJ25 isoform X2 [Jatropha curcas] |
| 19 |
Hb_002521_020 |
0.0794029363 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g03100, mitochondrial-like isoform X2 [Jatropha curcas] |
| 20 |
Hb_000363_300 |
0.0794614727 |
- |
- |
PREDICTED: protein ENHANCED DOWNY MILDEW 2 isoform X1 [Jatropha curcas] |