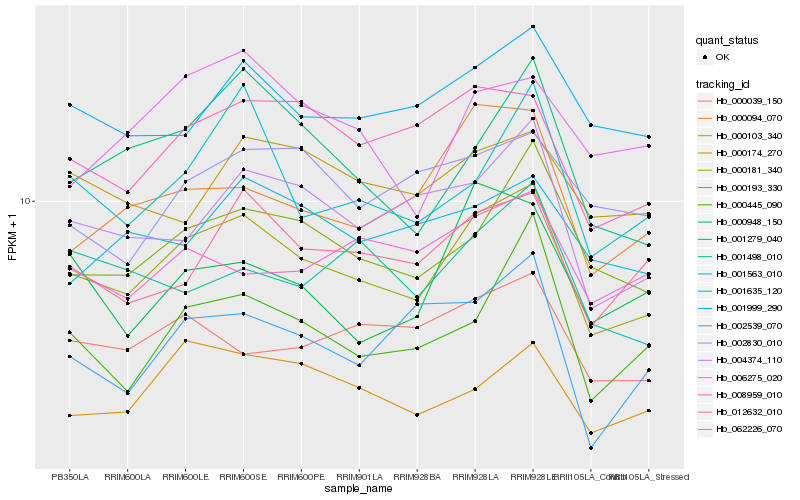
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000181_340 |
0.0 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g54610 [Jatropha curcas] |
| 2 |
Hb_000193_330 |
0.0684102552 |
- |
- |
PREDICTED: formin-like protein 20 [Jatropha curcas] |
| 3 |
Hb_012632_010 |
0.0721507568 |
- |
- |
PREDICTED: uncharacterized protein LOC105640019 [Jatropha curcas] |
| 4 |
Hb_062226_070 |
0.0750811566 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 4 isoform X1 [Jatropha curcas] |
| 5 |
Hb_004374_110 |
0.0752976436 |
- |
- |
PREDICTED: uncharacterized protein LOC105645768 [Jatropha curcas] |
| 6 |
Hb_000103_340 |
0.0767818778 |
- |
- |
PREDICTED: uncharacterized protein LOC105632051 isoform X4 [Jatropha curcas] |
| 7 |
Hb_001999_290 |
0.0776728451 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FTSH 7, chloroplastic-like [Jatropha curcas] |
| 8 |
Hb_001279_040 |
0.0777902244 |
- |
- |
PREDICTED: probable carboxylesterase 11 [Jatropha curcas] |
| 9 |
Hb_000445_090 |
0.0792093603 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase ASHH2 [Jatropha curcas] |
| 10 |
Hb_001563_010 |
0.0813548074 |
transcription factor |
TF Family: SET |
PREDICTED: probable histone-lysine N-methyltransferase ATXR3 [Jatropha curcas] |
| 11 |
Hb_000094_070 |
0.0823532819 |
- |
- |
PREDICTED: 5'-nucleotidase domain-containing protein 4 isoform X2 [Jatropha curcas] |
| 12 |
Hb_000039_150 |
0.0860650325 |
- |
- |
PREDICTED: lysine-specific demethylase JMJ25 isoform X1 [Jatropha curcas] |
| 13 |
Hb_001635_120 |
0.0875761274 |
- |
- |
PREDICTED: uncharacterized protein At1g51745-like [Jatropha curcas] |
| 14 |
Hb_000174_270 |
0.0875828989 |
- |
- |
PREDICTED: type II inositol 1,4,5-trisphosphate 5-phosphatase FRA3 isoform X1 [Jatropha curcas] |
| 15 |
Hb_006275_020 |
0.0884549672 |
- |
- |
PREDICTED: VIN3-like protein 2 [Jatropha curcas] |
| 16 |
Hb_002539_070 |
0.08883605 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g26460, mitochondrial [Jatropha curcas] |
| 17 |
Hb_008959_010 |
0.0910590956 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_001498_010 |
0.0923043547 |
- |
- |
PREDICTED: uncharacterized protein LOC105648994 [Jatropha curcas] |
| 19 |
Hb_002830_010 |
0.0924009615 |
- |
- |
PREDICTED: TBCC domain-containing protein 1 [Jatropha curcas] |
| 20 |
Hb_000948_150 |
0.0924244858 |
- |
- |
PREDICTED: heme-binding-like protein At3g10130, chloroplastic [Jatropha curcas] |