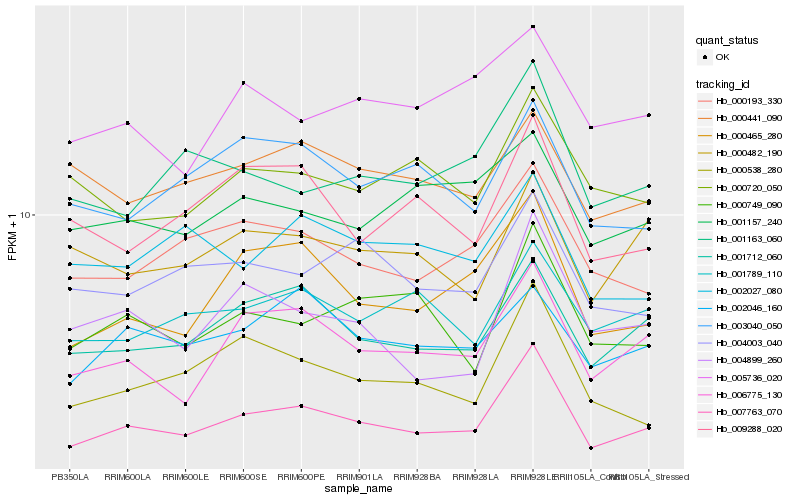
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007763_070 |
0.0 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g71460, chloroplastic [Jatropha curcas] |
| 2 |
Hb_001712_060 |
0.0641940878 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK9 [Jatropha curcas] |
| 3 |
Hb_006775_130 |
0.0654955009 |
- |
- |
PREDICTED: bidirectional sugar transporter SWEET2a-like [Jatropha curcas] |
| 4 |
Hb_000193_330 |
0.0712007741 |
- |
- |
PREDICTED: formin-like protein 20 [Jatropha curcas] |
| 5 |
Hb_000465_280 |
0.0747265622 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g47360 [Jatropha curcas] |
| 6 |
Hb_001789_110 |
0.0769590633 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 7 |
Hb_000538_280 |
0.0772230751 |
- |
- |
PREDICTED: uncharacterized protein LOC105642112 [Jatropha curcas] |
| 8 |
Hb_004003_040 |
0.0784830661 |
- |
- |
dead box ATP-dependent RNA helicase, putative [Ricinus communis] |
| 9 |
Hb_001163_060 |
0.0795385459 |
- |
- |
neutral/alkaline invertase [Manihot esculenta] |
| 10 |
Hb_000749_090 |
0.0803731209 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 11 |
Hb_002046_160 |
0.0818964504 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 12 |
Hb_000720_050 |
0.0831933733 |
- |
- |
PREDICTED: uncharacterized protein LOC105645857 isoform X1 [Jatropha curcas] |
| 13 |
Hb_001157_240 |
0.0845380911 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 37-like [Jatropha curcas] |
| 14 |
Hb_000482_190 |
0.0846249058 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1 [Jatropha curcas] |
| 15 |
Hb_004899_260 |
0.085284653 |
- |
- |
PREDICTED: uncharacterized protein LOC105638006 [Jatropha curcas] |
| 16 |
Hb_003040_050 |
0.0856437197 |
- |
- |
PREDICTED: uncharacterized protein LOC105638219 [Jatropha curcas] |
| 17 |
Hb_000441_090 |
0.0857447729 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 6 regulatory subunit 2 [Jatropha curcas] |
| 18 |
Hb_009288_020 |
0.0861471655 |
- |
- |
PREDICTED: protein FRIGIDA [Jatropha curcas] |
| 19 |
Hb_002027_080 |
0.0882544695 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105650047 [Jatropha curcas] |
| 20 |
Hb_005736_020 |
0.0884312348 |
- |
- |
hypothetical protein POPTR_0005s20940g [Populus trichocarpa] |