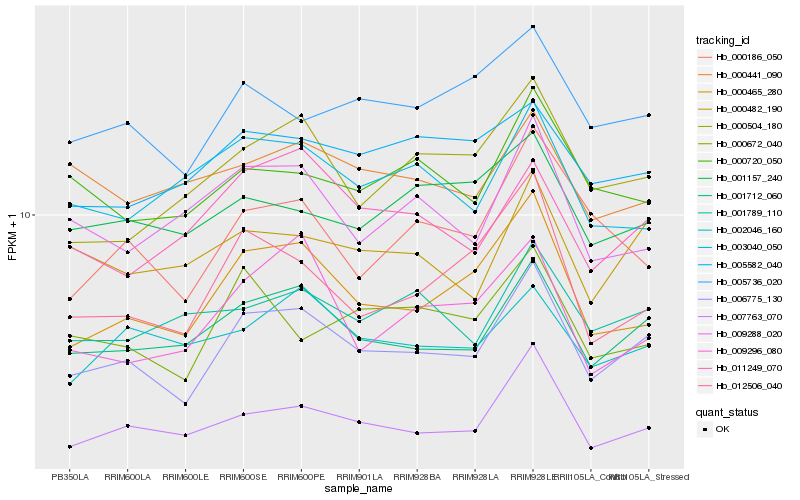
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006775_130 |
0.0 |
- |
- |
PREDICTED: bidirectional sugar transporter SWEET2a-like [Jatropha curcas] |
| 2 |
Hb_001712_060 |
0.0576975061 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK9 [Jatropha curcas] |
| 3 |
Hb_007763_070 |
0.0654955009 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g71460, chloroplastic [Jatropha curcas] |
| 4 |
Hb_000465_280 |
0.0706262764 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g47360 [Jatropha curcas] |
| 5 |
Hb_005736_020 |
0.0859613776 |
- |
- |
hypothetical protein POPTR_0005s20940g [Populus trichocarpa] |
| 6 |
Hb_000482_190 |
0.086966501 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1 [Jatropha curcas] |
| 7 |
Hb_000504_180 |
0.0883882992 |
- |
- |
PREDICTED: heterogeneous nuclear ribonucleoprotein 1 [Jatropha curcas] |
| 8 |
Hb_012506_040 |
0.0885104983 |
transcription factor |
TF Family: DDT |
PREDICTED: uncharacterized protein LOC105637346 [Jatropha curcas] |
| 9 |
Hb_011249_070 |
0.0913605311 |
- |
- |
hypothetical protein POPTR_0018s12610g [Populus trichocarpa] |
| 10 |
Hb_002046_160 |
0.0916877513 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 11 |
Hb_001789_110 |
0.092158628 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 12 |
Hb_000720_050 |
0.0923336841 |
- |
- |
PREDICTED: uncharacterized protein LOC105645857 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000672_040 |
0.092810398 |
- |
- |
PREDICTED: ENHANCER OF AG-4 protein 2 [Jatropha curcas] |
| 14 |
Hb_000186_050 |
0.0936757454 |
- |
- |
PREDICTED: uncharacterized protein LOC105649281 [Jatropha curcas] |
| 15 |
Hb_005582_040 |
0.0942173207 |
- |
- |
PREDICTED: regulator of nonsense transcripts 1 homolog [Jatropha curcas] |
| 16 |
Hb_009288_020 |
0.0954266941 |
- |
- |
PREDICTED: protein FRIGIDA [Jatropha curcas] |
| 17 |
Hb_009296_080 |
0.0964615189 |
- |
- |
PREDICTED: probable apyrase 7 isoform X1 [Jatropha curcas] |
| 18 |
Hb_001157_240 |
0.0967007437 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 37-like [Jatropha curcas] |
| 19 |
Hb_000441_090 |
0.0991738507 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 6 regulatory subunit 2 [Jatropha curcas] |
| 20 |
Hb_003040_050 |
0.099249017 |
- |
- |
PREDICTED: uncharacterized protein LOC105638219 [Jatropha curcas] |