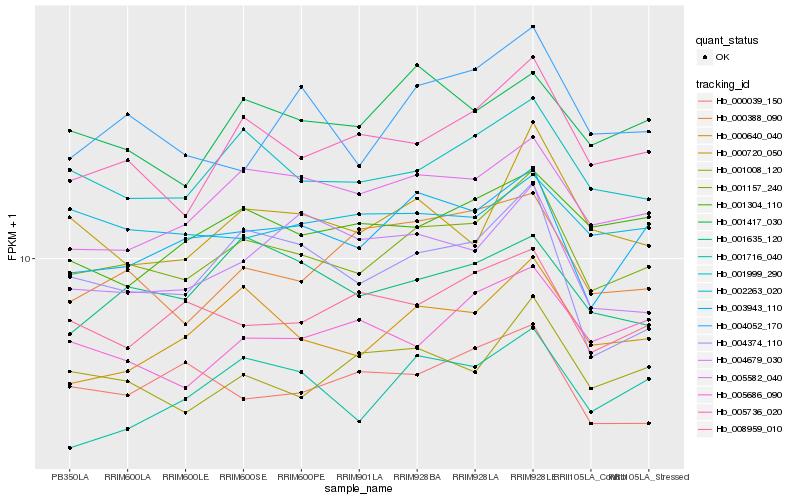
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001157_240 |
0.0 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 37-like [Jatropha curcas] |
| 2 |
Hb_001999_290 |
0.0521859762 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FTSH 7, chloroplastic-like [Jatropha curcas] |
| 3 |
Hb_003943_110 |
0.0553660481 |
transcription factor |
TF Family: MYB-related |
Zuotin, putative [Ricinus communis] |
| 4 |
Hb_005736_020 |
0.0554805203 |
- |
- |
hypothetical protein POPTR_0005s20940g [Populus trichocarpa] |
| 5 |
Hb_005582_040 |
0.0608999376 |
- |
- |
PREDICTED: regulator of nonsense transcripts 1 homolog [Jatropha curcas] |
| 6 |
Hb_000640_040 |
0.0613518754 |
- |
- |
PREDICTED: bromodomain and WD repeat-containing protein 1 isoform X2 [Jatropha curcas] |
| 7 |
Hb_001008_120 |
0.0668069676 |
desease resistance |
Gene Name: PEX-1N |
peroxisome biogenesis factor, putative [Ricinus communis] |
| 8 |
Hb_001417_030 |
0.0679437003 |
- |
- |
PREDICTED: inactive poly [ADP-ribose] polymerase RCD1 [Jatropha curcas] |
| 9 |
Hb_004374_110 |
0.0695659236 |
- |
- |
PREDICTED: uncharacterized protein LOC105645768 [Jatropha curcas] |
| 10 |
Hb_008959_010 |
0.0705272164 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000720_050 |
0.0713453436 |
- |
- |
PREDICTED: uncharacterized protein LOC105645857 isoform X1 [Jatropha curcas] |
| 12 |
Hb_001716_040 |
0.0716604919 |
- |
- |
ribonuclease p/mrp subunit, putative [Ricinus communis] |
| 13 |
Hb_001635_120 |
0.0722033887 |
- |
- |
PREDICTED: uncharacterized protein At1g51745-like [Jatropha curcas] |
| 14 |
Hb_000388_090 |
0.0737922772 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g77360, mitochondrial-like [Jatropha curcas] |
| 15 |
Hb_001304_110 |
0.0738687987 |
transcription factor |
TF Family: WRKY |
WRKY protein [Hevea brasiliensis] |
| 16 |
Hb_004052_170 |
0.0739738111 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein PNM1, mitochondrial [Jatropha curcas] |
| 17 |
Hb_002263_020 |
0.0751108993 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 11-like [Jatropha curcas] |
| 18 |
Hb_005686_090 |
0.0752760641 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 2 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000039_150 |
0.0776671647 |
- |
- |
PREDICTED: lysine-specific demethylase JMJ25 isoform X1 [Jatropha curcas] |
| 20 |
Hb_004679_030 |
0.0780357607 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: F-box protein SKIP23-like [Jatropha curcas] |