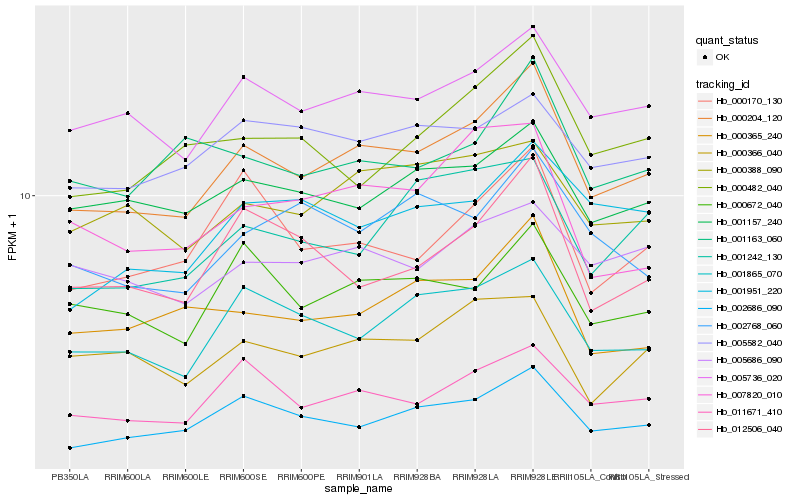
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000204_120 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105639239 [Jatropha curcas] |
| 2 |
Hb_005736_020 |
0.0644044992 |
- |
- |
hypothetical protein POPTR_0005s20940g [Populus trichocarpa] |
| 3 |
Hb_002686_090 |
0.0692407027 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g13650 [Jatropha curcas] |
| 4 |
Hb_011671_410 |
0.0709936539 |
transcription factor |
TF Family: SET |
histone-lysine n-methyltransferase, suvh, putative [Ricinus communis] |
| 5 |
Hb_001865_070 |
0.0801128638 |
- |
- |
PREDICTED: uncharacterized protein LOC105647937 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000388_090 |
0.0844130026 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g77360, mitochondrial-like [Jatropha curcas] |
| 7 |
Hb_000672_040 |
0.0847592889 |
- |
- |
PREDICTED: ENHANCER OF AG-4 protein 2 [Jatropha curcas] |
| 8 |
Hb_001157_240 |
0.0849390304 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 37-like [Jatropha curcas] |
| 9 |
Hb_000365_240 |
0.0880889801 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 33A-like [Jatropha curcas] |
| 10 |
Hb_005686_090 |
0.0890293421 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 2 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000482_040 |
0.089294051 |
- |
- |
Protein YME1, putative [Ricinus communis] |
| 12 |
Hb_000170_130 |
0.0894243512 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase ASHH2 [Jatropha curcas] |
| 13 |
Hb_012506_040 |
0.0903559709 |
transcription factor |
TF Family: DDT |
PREDICTED: uncharacterized protein LOC105637346 [Jatropha curcas] |
| 14 |
Hb_000366_040 |
0.0917764093 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g66520-like [Jatropha curcas] |
| 15 |
Hb_005582_040 |
0.0925571526 |
- |
- |
PREDICTED: regulator of nonsense transcripts 1 homolog [Jatropha curcas] |
| 16 |
Hb_001242_130 |
0.0929363107 |
- |
- |
PREDICTED: U3 small nucleolar RNA-associated protein 4-like [Jatropha curcas] |
| 17 |
Hb_001163_060 |
0.0930089654 |
- |
- |
neutral/alkaline invertase [Manihot esculenta] |
| 18 |
Hb_001951_220 |
0.0939084426 |
- |
- |
PREDICTED: CTD small phosphatase-like protein 2 isoform X5 [Jatropha curcas] |
| 19 |
Hb_002768_060 |
0.094302484 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 2 isoform X1 [Jatropha curcas] |
| 20 |
Hb_007820_010 |
0.0944188055 |
transcription factor |
TF Family: SWI/SNF-SWI3 |
PREDICTED: protein FLOWERING LOCUS D [Jatropha curcas] |