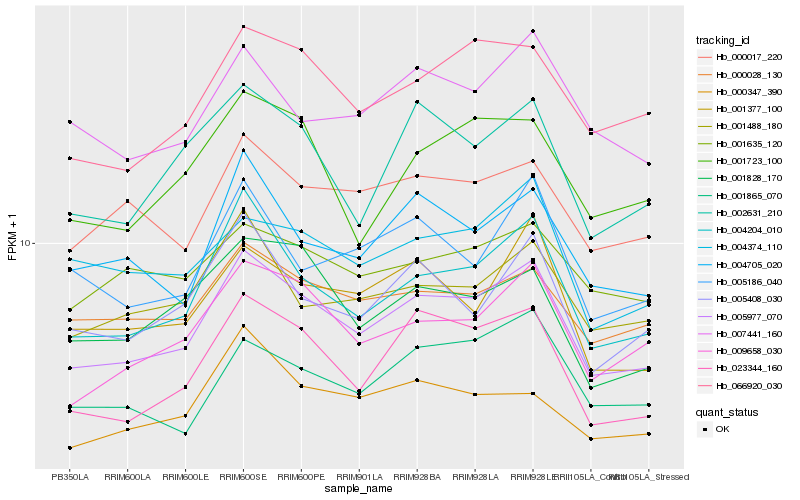
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005977_070 |
0.0 |
- |
- |
PREDICTED: probable helicase MAGATAMA 3 isoform X1 [Jatropha curcas] |
| 2 |
Hb_004204_010 |
0.0503941401 |
- |
- |
PREDICTED: uncharacterized protein LOC105640608 [Jatropha curcas] |
| 3 |
Hb_023344_160 |
0.0588526129 |
- |
- |
PREDICTED: uncharacterized protein LOC105644462 [Jatropha curcas] |
| 4 |
Hb_009658_030 |
0.063480405 |
- |
- |
PREDICTED: AP-5 complex subunit beta-1 [Jatropha curcas] |
| 5 |
Hb_066920_030 |
0.0676621241 |
- |
- |
PREDICTED: adagio protein 1 [Jatropha curcas] |
| 6 |
Hb_004705_020 |
0.0711506988 |
- |
- |
PREDICTED: RNA polymerase II C-terminal domain phosphatase-like 3 [Jatropha curcas] |
| 7 |
Hb_000017_220 |
0.071191708 |
- |
- |
PREDICTED: uncharacterized protein LOC105645768 [Jatropha curcas] |
| 8 |
Hb_005408_030 |
0.0733194339 |
- |
- |
PREDICTED: uncharacterized protein LOC105648043 [Jatropha curcas] |
| 9 |
Hb_001488_180 |
0.0735296759 |
- |
- |
PREDICTED: uncharacterized protein LOC105644152 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000028_130 |
0.0739609243 |
- |
- |
PREDICTED: uncharacterized protein LOC105645303 isoform X1 [Jatropha curcas] |
| 11 |
Hb_001865_070 |
0.0744280455 |
- |
- |
PREDICTED: uncharacterized protein LOC105647937 isoform X1 [Jatropha curcas] |
| 12 |
Hb_001377_100 |
0.0744847386 |
- |
- |
PREDICTED: uncharacterized protein LOC105642863 isoform X1 [Jatropha curcas] |
| 13 |
Hb_002631_210 |
0.0756309175 |
- |
- |
PREDICTED: uncharacterized protein LOC105646172 [Jatropha curcas] |
| 14 |
Hb_007441_160 |
0.0761901802 |
- |
- |
RNA binding protein, putative [Ricinus communis] |
| 15 |
Hb_001828_170 |
0.0778071347 |
- |
- |
PREDICTED: protein TPLATE [Jatropha curcas] |
| 16 |
Hb_004374_110 |
0.0779036664 |
- |
- |
PREDICTED: uncharacterized protein LOC105645768 [Jatropha curcas] |
| 17 |
Hb_000347_390 |
0.0790053173 |
- |
- |
PREDICTED: exportin-T [Jatropha curcas] |
| 18 |
Hb_001723_100 |
0.0790190171 |
- |
- |
PREDICTED: uncharacterized protein LOC105643601 isoform X1 [Jatropha curcas] |
| 19 |
Hb_005186_040 |
0.0802639864 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 13 isoform X2 [Jatropha curcas] |
| 20 |
Hb_001635_120 |
0.0832602542 |
- |
- |
PREDICTED: uncharacterized protein At1g51745-like [Jatropha curcas] |