| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
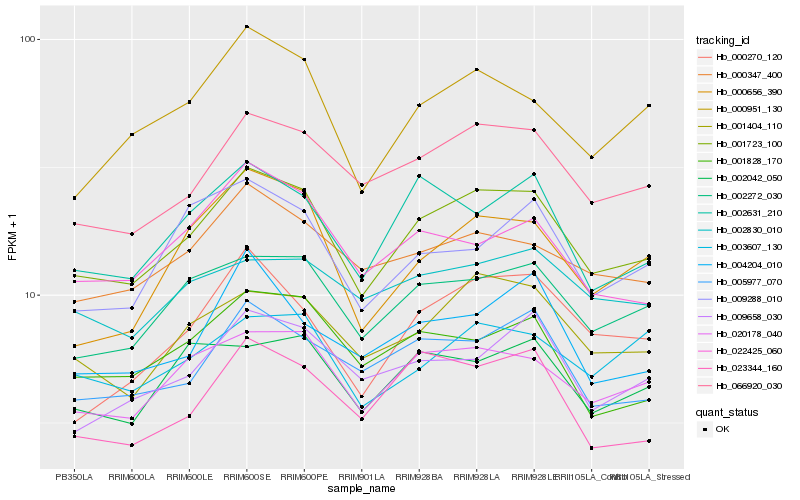
Hb_001723_100 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105643601 isoform X1 [Jatropha curcas] |
| 2 |
Hb_020178_040 |
0.0605115399 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_066920_030 |
0.0610324897 |
- |
- |
PREDICTED: adagio protein 1 [Jatropha curcas] |
| 4 |
Hb_002272_030 |
0.0638992102 |
- |
- |
PREDICTED: cation/calcium exchanger 5 [Jatropha curcas] |
| 5 |
Hb_001404_110 |
0.067221122 |
- |
- |
phosphoinositide 5-phosphatase, putative [Ricinus communis] |
| 6 |
Hb_002631_210 |
0.0760719709 |
- |
- |
PREDICTED: uncharacterized protein LOC105646172 [Jatropha curcas] |
| 7 |
Hb_000347_400 |
0.0788144221 |
- |
- |
PREDICTED: probable GTP diphosphokinase RSH2, chloroplastic [Jatropha curcas] |
| 8 |
Hb_005977_070 |
0.0790190171 |
- |
- |
PREDICTED: probable helicase MAGATAMA 3 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000951_130 |
0.0800669995 |
- |
- |
PREDICTED: transcription factor GTE2-like [Jatropha curcas] |
| 10 |
Hb_002042_050 |
0.084560835 |
- |
- |
PREDICTED: uncharacterized protein LOC105647554 isoform X1 [Jatropha curcas] |
| 11 |
Hb_009658_030 |
0.085278371 |
- |
- |
PREDICTED: AP-5 complex subunit beta-1 [Jatropha curcas] |
| 12 |
Hb_001828_170 |
0.0868918814 |
- |
- |
PREDICTED: protein TPLATE [Jatropha curcas] |
| 13 |
Hb_000656_390 |
0.0870128491 |
- |
- |
PREDICTED: probable GTP diphosphokinase RSH2, chloroplastic [Jatropha curcas] |
| 14 |
Hb_023344_160 |
0.0871182361 |
- |
- |
PREDICTED: uncharacterized protein LOC105644462 [Jatropha curcas] |
| 15 |
Hb_000270_120 |
0.0871934365 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g18900 [Jatropha curcas] |
| 16 |
Hb_022425_060 |
0.0871992243 |
- |
- |
PREDICTED: protein phosphatase 2C and cyclic nucleotide-binding/kinase domain-containing protein isoform X1 [Jatropha curcas] |
| 17 |
Hb_003607_130 |
0.0892702961 |
- |
- |
serine/threonine protein kinase, putative [Ricinus communis] |
| 18 |
Hb_002830_010 |
0.0895782452 |
- |
- |
PREDICTED: TBCC domain-containing protein 1 [Jatropha curcas] |
| 19 |
Hb_004204_010 |
0.0909560071 |
- |
- |
PREDICTED: uncharacterized protein LOC105640608 [Jatropha curcas] |
| 20 |
Hb_009288_010 |
0.0910075322 |
desease resistance |
Gene Name: Clp_N |
ERD1 protein, chloroplast precursor, putative [Ricinus communis] |