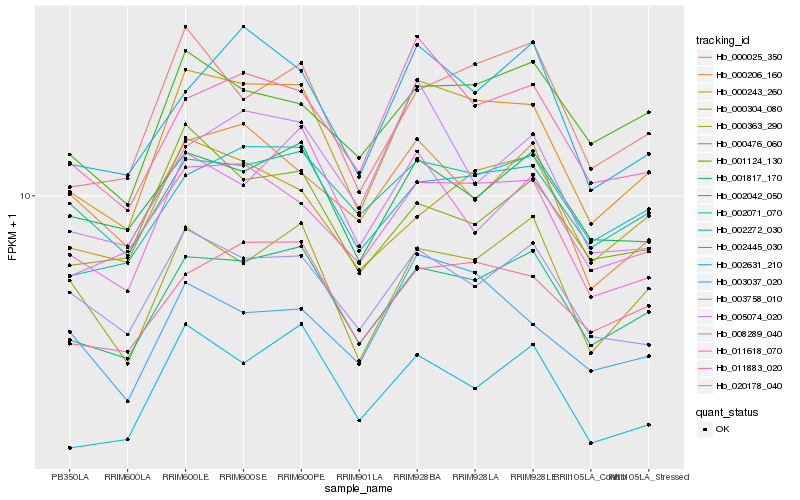
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000243_260 |
0.0 |
- |
- |
PREDICTED: UPF0183 protein At3g51130 isoform X2 [Jatropha curcas] |
| 2 |
Hb_002042_050 |
0.0569452305 |
- |
- |
PREDICTED: uncharacterized protein LOC105647554 isoform X1 [Jatropha curcas] |
| 3 |
Hb_011618_070 |
0.0714654308 |
- |
- |
NMDA receptor-regulated protein [Medicago truncatula] |
| 4 |
Hb_020178_040 |
0.0727184439 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_011883_020 |
0.0748027064 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: aspartate--tRNA ligase, cytoplasmic-like [Jatropha curcas] |
| 6 |
Hb_003758_010 |
0.0820197101 |
transcription factor |
TF Family: VOZ |
PREDICTED: transcription factor VOZ1 isoform X1 [Jatropha curcas] |
| 7 |
Hb_002631_210 |
0.0822975566 |
- |
- |
PREDICTED: uncharacterized protein LOC105646172 [Jatropha curcas] |
| 8 |
Hb_002272_030 |
0.0823211976 |
- |
- |
PREDICTED: cation/calcium exchanger 5 [Jatropha curcas] |
| 9 |
Hb_000025_350 |
0.0823818516 |
- |
- |
PREDICTED: rho GTPase-activating protein 7 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000363_290 |
0.0843028453 |
- |
- |
fimbrin, putative [Ricinus communis] |
| 11 |
Hb_008289_040 |
0.0855924735 |
- |
- |
PREDICTED: suppressor of mec-8 and unc-52 protein homolog 1 [Jatropha curcas] |
| 12 |
Hb_002071_070 |
0.0857636078 |
transcription factor |
TF Family: RB |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_001817_170 |
0.0870669471 |
- |
- |
PREDICTED: AP-1 complex subunit gamma-2-like [Jatropha curcas] |
| 14 |
Hb_002445_030 |
0.0878374289 |
- |
- |
hypothetical protein JCGZ_12656 [Jatropha curcas] |
| 15 |
Hb_000476_060 |
0.0901322374 |
- |
- |
Tetratricopeptide repeat (TPR)-like superfamily protein isoform 1 [Theobroma cacao] |
| 16 |
Hb_000304_080 |
0.0918087931 |
- |
- |
PREDICTED: probable helicase MAGATAMA 3 isoform X1 [Jatropha curcas] |
| 17 |
Hb_001124_130 |
0.092018505 |
transcription factor |
TF Family: TUB |
phosphoric diester hydrolase, putative [Ricinus communis] |
| 18 |
Hb_003037_020 |
0.0921458841 |
- |
- |
hypothetical protein JCGZ_22910 [Jatropha curcas] |
| 19 |
Hb_000206_160 |
0.0924895988 |
- |
- |
PREDICTED: TBCC domain-containing protein 1 [Jatropha curcas] |
| 20 |
Hb_005074_020 |
0.0942243023 |
- |
- |
PREDICTED: uncharacterized protein LOC105644455 isoform X1 [Jatropha curcas] |