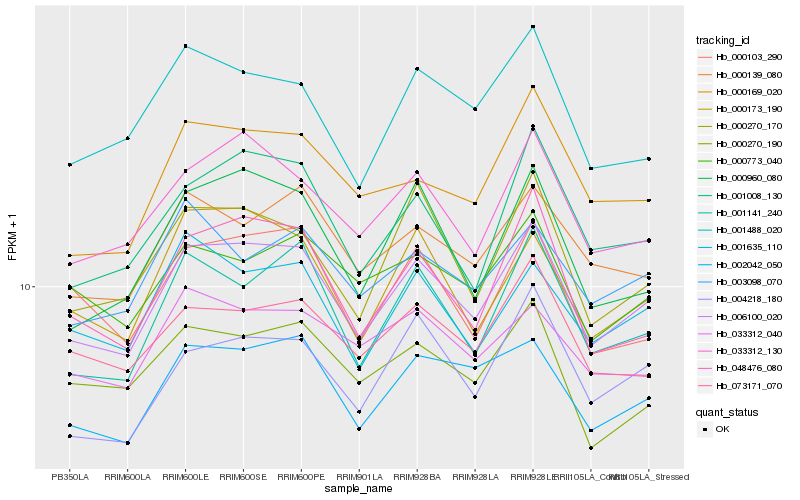
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006100_020 |
0.0 |
- |
- |
PREDICTED: inactive poly [ADP-ribose] polymerase RCD1 [Jatropha curcas] |
| 2 |
Hb_002042_050 |
0.0572473459 |
- |
- |
PREDICTED: uncharacterized protein LOC105647554 isoform X1 [Jatropha curcas] |
| 3 |
Hb_033312_130 |
0.0573539727 |
- |
- |
PREDICTED: protein TIC 40, chloroplastic [Jatropha curcas] |
| 4 |
Hb_001141_240 |
0.0584803561 |
- |
- |
PREDICTED: folate-biopterin transporter 1, chloroplastic [Jatropha curcas] |
| 5 |
Hb_048476_080 |
0.0617432893 |
transcription factor |
TF Family: ARR-B |
two-component sensor histidine kinase bacteria, putative [Ricinus communis] |
| 6 |
Hb_001008_130 |
0.0618920344 |
transcription factor |
TF Family: mTERF |
PREDICTED: cytochrome c1-2, heme protein, mitochondrial-like [Elaeis guineensis] |
| 7 |
Hb_000169_020 |
0.0636539369 |
transcription factor |
TF Family: C2C2-CO-like |
hypothetical protein RCOM_0555710 [Ricinus communis] |
| 8 |
Hb_000139_080 |
0.0637918026 |
- |
- |
PREDICTED: vacuole membrane protein KMS1 isoform X2 [Jatropha curcas] |
| 9 |
Hb_000173_190 |
0.0639005006 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000270_170 |
0.0646225108 |
- |
- |
PREDICTED: formin-like protein 5 [Jatropha curcas] |
| 11 |
Hb_004218_180 |
0.0653531241 |
- |
- |
PREDICTED: flowering time control protein FY isoform X2 [Jatropha curcas] |
| 12 |
Hb_001635_110 |
0.0661049911 |
- |
- |
PREDICTED: NADP-specific glutamate dehydrogenase [Jatropha curcas] |
| 13 |
Hb_073171_070 |
0.0671235478 |
- |
- |
PREDICTED: RNA polymerase II-associated factor 1 homolog [Jatropha curcas] |
| 14 |
Hb_001488_020 |
0.0676313399 |
- |
- |
PREDICTED: probable mitochondrial-processing peptidase subunit beta [Jatropha curcas] |
| 15 |
Hb_000960_080 |
0.0686966777 |
- |
- |
PREDICTED: topless-related protein 1-like isoform X2 [Jatropha curcas] |
| 16 |
Hb_000103_290 |
0.0704890377 |
- |
- |
PREDICTED: dymeclin isoform X1 [Jatropha curcas] |
| 17 |
Hb_033312_040 |
0.0722328656 |
- |
- |
PREDICTED: gamma-tubulin complex component 5-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_000773_040 |
0.072445573 |
- |
- |
PREDICTED: uncharacterized protein LOC105641863 isoform X2 [Jatropha curcas] |
| 19 |
Hb_000270_190 |
0.0726684811 |
transcription factor |
TF Family: DDT |
tRNA ligase, putative [Ricinus communis] |
| 20 |
Hb_003098_070 |
0.0731709516 |
- |
- |
PREDICTED: uncharacterized protein LOC105633456 [Jatropha curcas] |