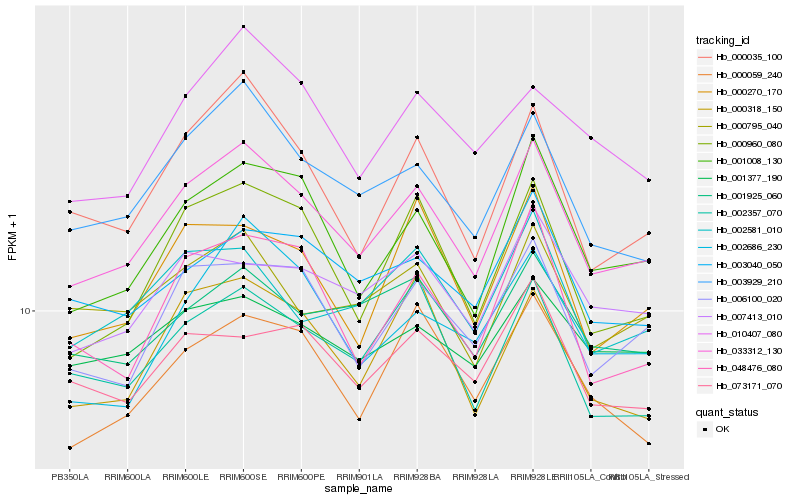
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_033312_130 |
0.0 |
- |
- |
PREDICTED: protein TIC 40, chloroplastic [Jatropha curcas] |
| 2 |
Hb_003929_210 |
0.0520787202 |
transcription factor |
TF Family: SBP |
Squamosa promoter-binding protein, putative [Ricinus communis] |
| 3 |
Hb_001008_130 |
0.054191011 |
transcription factor |
TF Family: mTERF |
PREDICTED: cytochrome c1-2, heme protein, mitochondrial-like [Elaeis guineensis] |
| 4 |
Hb_006100_020 |
0.0573539727 |
- |
- |
PREDICTED: inactive poly [ADP-ribose] polymerase RCD1 [Jatropha curcas] |
| 5 |
Hb_000960_080 |
0.0626059796 |
- |
- |
PREDICTED: topless-related protein 1-like isoform X2 [Jatropha curcas] |
| 6 |
Hb_000035_100 |
0.0636342691 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 7 |
Hb_002357_070 |
0.0663942345 |
transcription factor |
TF Family: PHD |
PREDICTED: protein strawberry notch homolog 1 [Jatropha curcas] |
| 8 |
Hb_048476_080 |
0.0673312681 |
transcription factor |
TF Family: ARR-B |
two-component sensor histidine kinase bacteria, putative [Ricinus communis] |
| 9 |
Hb_000318_150 |
0.0678674756 |
- |
- |
RNA-binding protein Nova-1, putative [Ricinus communis] |
| 10 |
Hb_003040_050 |
0.0681856161 |
- |
- |
PREDICTED: uncharacterized protein LOC105638219 [Jatropha curcas] |
| 11 |
Hb_000059_240 |
0.0689443417 |
- |
- |
PREDICTED: protein FRIGIDA [Jatropha curcas] |
| 12 |
Hb_007413_010 |
0.0691337038 |
- |
- |
PREDICTED: SUMO-activating enzyme subunit 2 [Jatropha curcas] |
| 13 |
Hb_000270_170 |
0.0695852945 |
- |
- |
PREDICTED: formin-like protein 5 [Jatropha curcas] |
| 14 |
Hb_002581_010 |
0.0702312521 |
- |
- |
PREDICTED: uncharacterized protein LOC105641469 isoform X2 [Jatropha curcas] |
| 15 |
Hb_001925_060 |
0.0707626895 |
- |
- |
PREDICTED: putative RNA polymerase II subunit B1 CTD phosphatase RPAP2 homolog [Jatropha curcas] |
| 16 |
Hb_000795_040 |
0.0715405188 |
- |
- |
PREDICTED: sister chromatid cohesion protein PDS5 homolog A isoform X1 [Jatropha curcas] |
| 17 |
Hb_010407_080 |
0.0735626155 |
- |
- |
PREDICTED: putative E3 ubiquitin-protein ligase XBAT35 isoform X1 [Jatropha curcas] |
| 18 |
Hb_002686_230 |
0.0736213376 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 19 |
Hb_073171_070 |
0.0736340064 |
- |
- |
PREDICTED: RNA polymerase II-associated factor 1 homolog [Jatropha curcas] |
| 20 |
Hb_001377_190 |
0.0740525399 |
transcription factor |
TF Family: PHD |
PREDICTED: histone-lysine N-methyltransferase ATX3 isoform X2 [Jatropha curcas] |