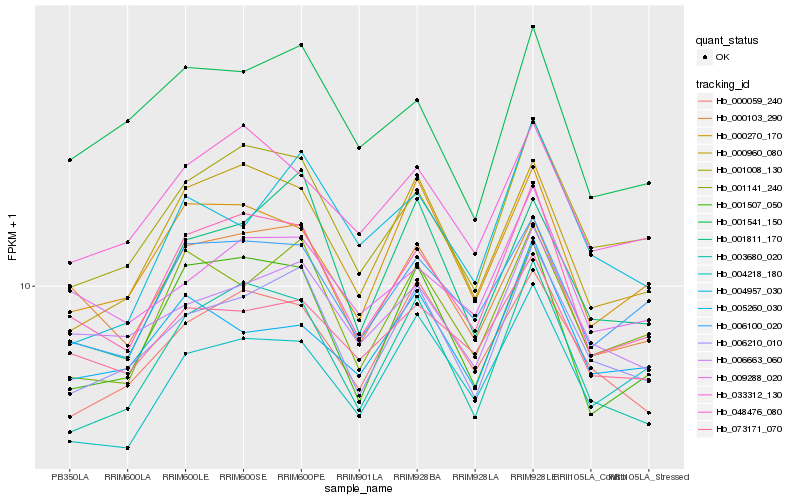
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006210_010 |
0.0 |
- |
- |
PREDICTED: probable acyl-activating enzyme 17, peroxisomal isoform X2 [Jatropha curcas] |
| 2 |
Hb_001008_130 |
0.0575047833 |
transcription factor |
TF Family: mTERF |
PREDICTED: cytochrome c1-2, heme protein, mitochondrial-like [Elaeis guineensis] |
| 3 |
Hb_006663_060 |
0.0601072715 |
- |
- |
PREDICTED: calcium homeostasis endoplasmic reticulum protein [Jatropha curcas] |
| 4 |
Hb_000059_240 |
0.0653953252 |
- |
- |
PREDICTED: protein FRIGIDA [Jatropha curcas] |
| 5 |
Hb_003680_020 |
0.0662757148 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 6 |
Hb_001141_240 |
0.0670720401 |
- |
- |
PREDICTED: folate-biopterin transporter 1, chloroplastic [Jatropha curcas] |
| 7 |
Hb_001811_170 |
0.0718980414 |
- |
- |
dynamin, putative [Ricinus communis] |
| 8 |
Hb_048476_080 |
0.0763584839 |
transcription factor |
TF Family: ARR-B |
two-component sensor histidine kinase bacteria, putative [Ricinus communis] |
| 9 |
Hb_000960_080 |
0.0765458919 |
- |
- |
PREDICTED: topless-related protein 1-like isoform X2 [Jatropha curcas] |
| 10 |
Hb_001541_150 |
0.0810063487 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000270_170 |
0.0817393977 |
- |
- |
PREDICTED: formin-like protein 5 [Jatropha curcas] |
| 12 |
Hb_006100_020 |
0.0824374237 |
- |
- |
PREDICTED: inactive poly [ADP-ribose] polymerase RCD1 [Jatropha curcas] |
| 13 |
Hb_073171_070 |
0.0829294549 |
- |
- |
PREDICTED: RNA polymerase II-associated factor 1 homolog [Jatropha curcas] |
| 14 |
Hb_000103_290 |
0.0838245498 |
- |
- |
PREDICTED: dymeclin isoform X1 [Jatropha curcas] |
| 15 |
Hb_033312_130 |
0.0841647607 |
- |
- |
PREDICTED: protein TIC 40, chloroplastic [Jatropha curcas] |
| 16 |
Hb_001507_050 |
0.0845282179 |
- |
- |
PREDICTED: cysteine desulfurase 1, chloroplastic isoform X1 [Jatropha curcas] |
| 17 |
Hb_005260_030 |
0.0857011175 |
- |
- |
PREDICTED: uncharacterized protein LOC105633782 [Jatropha curcas] |
| 18 |
Hb_009288_020 |
0.0859735225 |
- |
- |
PREDICTED: protein FRIGIDA [Jatropha curcas] |
| 19 |
Hb_004957_030 |
0.0863505355 |
- |
- |
hydroxyproline-rich glycoprotein [Populus trichocarpa] |
| 20 |
Hb_004218_180 |
0.0871402812 |
- |
- |
PREDICTED: flowering time control protein FY isoform X2 [Jatropha curcas] |