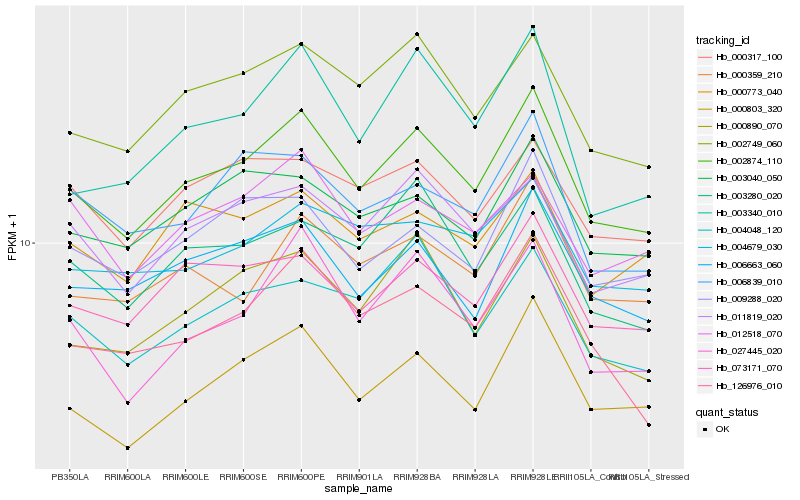
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002874_110 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105637451 [Jatropha curcas] |
| 2 |
Hb_002749_060 |
0.0568860203 |
- |
- |
PREDICTED: protein transport protein Sec61 subunit alpha-like [Jatropha curcas] |
| 3 |
Hb_000803_320 |
0.0575524159 |
transcription factor |
TF Family: PHD |
PREDICTED: histone-lysine N-methyltransferase ATX4 [Jatropha curcas] |
| 4 |
Hb_073171_070 |
0.061014001 |
- |
- |
PREDICTED: RNA polymerase II-associated factor 1 homolog [Jatropha curcas] |
| 5 |
Hb_006663_060 |
0.0644040544 |
- |
- |
PREDICTED: calcium homeostasis endoplasmic reticulum protein [Jatropha curcas] |
| 6 |
Hb_011819_020 |
0.0650180435 |
- |
- |
PREDICTED: uncharacterized protein LOC105643703 isoform X1 [Jatropha curcas] |
| 7 |
Hb_009288_020 |
0.0664270094 |
- |
- |
PREDICTED: protein FRIGIDA [Jatropha curcas] |
| 8 |
Hb_000359_210 |
0.066862158 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000890_070 |
0.0677489741 |
- |
- |
PREDICTED: protein ALWAYS EARLY 3 isoform X2 [Jatropha curcas] |
| 10 |
Hb_003040_050 |
0.0681649339 |
- |
- |
PREDICTED: uncharacterized protein LOC105638219 [Jatropha curcas] |
| 11 |
Hb_003280_020 |
0.069270321 |
- |
- |
PREDICTED: enhancer of mRNA-decapping protein 4 isoform X1 [Jatropha curcas] |
| 12 |
Hb_027445_020 |
0.069393587 |
- |
- |
PREDICTED: glycosylphosphatidylinositol anchor attachment 1 protein [Jatropha curcas] |
| 13 |
Hb_126976_010 |
0.0706487274 |
- |
- |
- |
| 14 |
Hb_004679_030 |
0.0721064711 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: F-box protein SKIP23-like [Jatropha curcas] |
| 15 |
Hb_003340_010 |
0.0721462285 |
- |
- |
PREDICTED: ubiquitin-activating enzyme E1 1 [Vitis vinifera] |
| 16 |
Hb_000317_100 |
0.0727515179 |
- |
- |
PREDICTED: uncharacterized protein LOC105646995 [Jatropha curcas] |
| 17 |
Hb_004048_120 |
0.0737800841 |
- |
- |
PREDICTED: nuclear pore complex protein NUP160 isoform X3 [Jatropha curcas] |
| 18 |
Hb_006839_010 |
0.0745271899 |
- |
- |
PREDICTED: importin subunit beta-1-like [Jatropha curcas] |
| 19 |
Hb_000773_040 |
0.0747513888 |
- |
- |
PREDICTED: uncharacterized protein LOC105641863 isoform X2 [Jatropha curcas] |
| 20 |
Hb_012518_070 |
0.0748666367 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 9 isoform X1 [Jatropha curcas] |