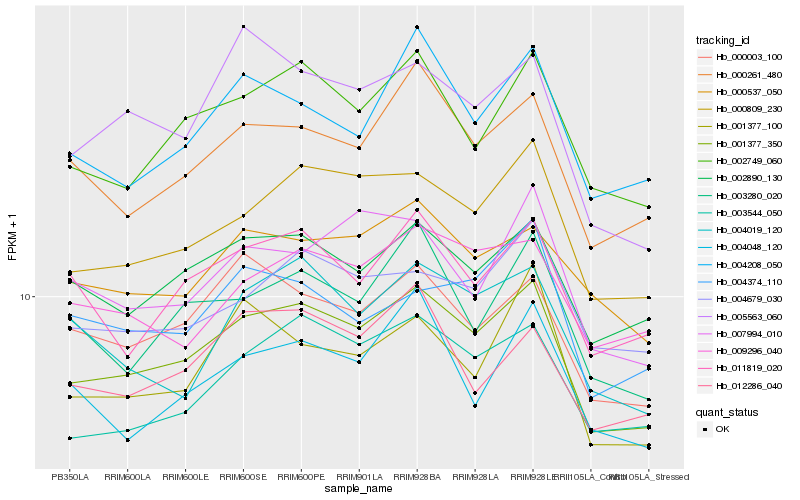
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001377_350 |
0.0 |
- |
- |
PREDICTED: exportin-2 [Jatropha curcas] |
| 2 |
Hb_000809_230 |
0.0473420006 |
- |
- |
PREDICTED: uncharacterized protein LOC105639681 isoform X1 [Jatropha curcas] |
| 3 |
Hb_002749_060 |
0.0533334376 |
- |
- |
PREDICTED: protein transport protein Sec61 subunit alpha-like [Jatropha curcas] |
| 4 |
Hb_002890_130 |
0.0558904534 |
- |
- |
tip120, putative [Ricinus communis] |
| 5 |
Hb_000261_480 |
0.0572877181 |
- |
- |
26S proteasome regulatory subunit family protein [Populus trichocarpa] |
| 6 |
Hb_003544_050 |
0.05920524 |
- |
- |
PREDICTED: uncharacterized protein LOC105644300 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000003_100 |
0.0618323466 |
transcription factor |
TF Family: SNF2 |
PREDICTED: transcription termination factor 2 isoform X5 [Jatropha curcas] |
| 8 |
Hb_004679_030 |
0.0645100287 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: F-box protein SKIP23-like [Jatropha curcas] |
| 9 |
Hb_011819_020 |
0.0666033425 |
- |
- |
PREDICTED: uncharacterized protein LOC105643703 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000537_050 |
0.0667366177 |
- |
- |
AP-2 complex subunit beta-1, putative [Ricinus communis] |
| 11 |
Hb_004208_050 |
0.0668067044 |
desease resistance |
Gene Name: CDC48_N |
cell division cycle protein 48 [Hevea brasiliensis] |
| 12 |
Hb_012286_040 |
0.0672530764 |
- |
- |
PREDICTED: pre-mRNA-splicing factor RSE1 [Jatropha curcas] |
| 13 |
Hb_003280_020 |
0.0679203014 |
- |
- |
PREDICTED: enhancer of mRNA-decapping protein 4 isoform X1 [Jatropha curcas] |
| 14 |
Hb_005563_060 |
0.0698434606 |
- |
- |
PREDICTED: coatomer subunit alpha-1 [Jatropha curcas] |
| 15 |
Hb_009296_040 |
0.0699764373 |
- |
- |
hypothetical protein L484_007435 [Morus notabilis] |
| 16 |
Hb_001377_100 |
0.070907413 |
- |
- |
PREDICTED: uncharacterized protein LOC105642863 isoform X1 [Jatropha curcas] |
| 17 |
Hb_004019_120 |
0.072663895 |
- |
- |
PREDICTED: conserved oligomeric Golgi complex subunit 1 [Jatropha curcas] |
| 18 |
Hb_007994_010 |
0.0731119576 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR-RED ELONGATED HYPOCOTYL 3 isoform X1 [Jatropha curcas] |
| 19 |
Hb_004048_120 |
0.0737733726 |
- |
- |
PREDICTED: nuclear pore complex protein NUP160 isoform X3 [Jatropha curcas] |
| 20 |
Hb_004374_110 |
0.0746014418 |
- |
- |
PREDICTED: uncharacterized protein LOC105645768 [Jatropha curcas] |