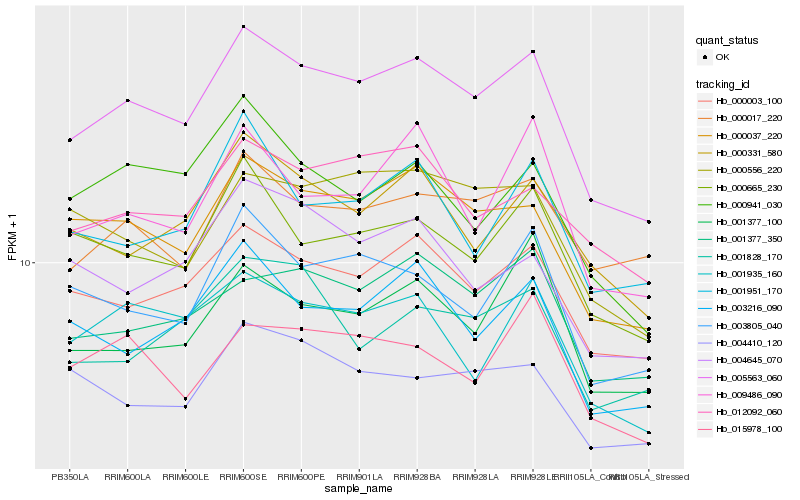
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005563_060 |
0.0 |
- |
- |
PREDICTED: coatomer subunit alpha-1 [Jatropha curcas] |
| 2 |
Hb_000037_220 |
0.0558711524 |
- |
- |
PREDICTED: vacuolar protein-sorting-associated protein 11 homolog [Jatropha curcas] |
| 3 |
Hb_000003_100 |
0.0569663576 |
transcription factor |
TF Family: SNF2 |
PREDICTED: transcription termination factor 2 isoform X5 [Jatropha curcas] |
| 4 |
Hb_001377_350 |
0.0698434606 |
- |
- |
PREDICTED: exportin-2 [Jatropha curcas] |
| 5 |
Hb_003216_090 |
0.072259484 |
- |
- |
PREDICTED: nuclear pore complex protein NUP133 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000665_230 |
0.0735226506 |
- |
- |
PREDICTED: uncharacterized protein LOC105637615 [Jatropha curcas] |
| 7 |
Hb_012092_060 |
0.0736379512 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375 [Jatropha curcas] |
| 8 |
Hb_000331_580 |
0.0747536485 |
- |
- |
PREDICTED: programmed cell death protein 4-like [Jatropha curcas] |
| 9 |
Hb_003805_040 |
0.0759561011 |
- |
- |
Sacsin [Glycine soja] |
| 10 |
Hb_001935_160 |
0.0769626791 |
- |
- |
PREDICTED: F-box protein At5g51370-like [Jatropha curcas] |
| 11 |
Hb_001377_100 |
0.0782759182 |
- |
- |
PREDICTED: uncharacterized protein LOC105642863 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000556_220 |
0.0790534872 |
- |
- |
PREDICTED: brefeldin A-inhibited guanine nucleotide-exchange protein 2-like [Jatropha curcas] |
| 13 |
Hb_009486_090 |
0.0805438507 |
- |
- |
RNA binding protein, putative [Ricinus communis] |
| 14 |
Hb_000017_220 |
0.0808115003 |
- |
- |
PREDICTED: uncharacterized protein LOC105645768 [Jatropha curcas] |
| 15 |
Hb_015978_100 |
0.0823800995 |
- |
- |
PREDICTED: DNA mismatch repair protein MSH6 [Jatropha curcas] |
| 16 |
Hb_001951_170 |
0.0832158535 |
- |
- |
PREDICTED: uncharacterized protein LOC105649755 [Jatropha curcas] |
| 17 |
Hb_004410_120 |
0.0833781922 |
- |
- |
PREDICTED: uncharacterized protein LOC105633877 isoform X1 [Jatropha curcas] |
| 18 |
Hb_004645_070 |
0.0836895389 |
- |
- |
PREDICTED: beta-1,3-galactosyltransferase 15 [Jatropha curcas] |
| 19 |
Hb_001828_170 |
0.0848292069 |
- |
- |
PREDICTED: protein TPLATE [Jatropha curcas] |
| 20 |
Hb_000941_030 |
0.0858831785 |
- |
- |
PREDICTED: uncharacterized protein LOC105646531 [Jatropha curcas] |