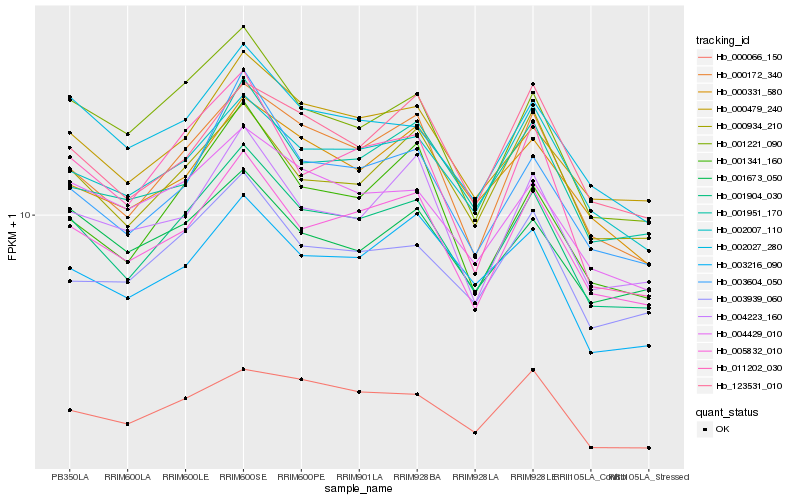
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001904_030 |
0.0 |
- |
- |
PREDICTED: importin-5 [Jatropha curcas] |
| 2 |
Hb_005832_010 |
0.0440085047 |
- |
- |
PREDICTED: enhancer of mRNA-decapping protein 4 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000172_340 |
0.0486851956 |
- |
- |
PREDICTED: uncharacterized protein LOC101261327 [Solanum lycopersicum] |
| 4 |
Hb_000479_240 |
0.0538476638 |
- |
- |
PREDICTED: probable ubiquitin conjugation factor E4 isoform X2 [Jatropha curcas] |
| 5 |
Hb_003604_050 |
0.0555536546 |
- |
- |
PREDICTED: MAP3K epsilon protein kinase 1-like isoform X2 [Jatropha curcas] |
| 6 |
Hb_002027_280 |
0.0607371901 |
- |
- |
PREDICTED: sister-chromatid cohesion protein 3 [Jatropha curcas] |
| 7 |
Hb_001221_090 |
0.0612886608 |
- |
- |
PREDICTED: uncharacterized protein LOC105640211 isoform X1 [Jatropha curcas] |
| 8 |
Hb_002007_110 |
0.0619418979 |
transcription factor |
TF Family: Jumonji |
PREDICTED: lysine-specific demethylase JMJ25 isoform X2 [Jatropha curcas] |
| 9 |
Hb_003939_060 |
0.0620786213 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_004429_010 |
0.0623855757 |
- |
- |
calcineurin-like phosphoesterase [Manihot esculenta] |
| 11 |
Hb_000934_210 |
0.0663938638 |
- |
- |
suppressor of ty, putative [Ricinus communis] |
| 12 |
Hb_004223_160 |
0.0671369711 |
- |
- |
PREDICTED: 5'-3' exoribonuclease 3 isoform X2 [Jatropha curcas] |
| 13 |
Hb_001951_170 |
0.0676771858 |
- |
- |
PREDICTED: uncharacterized protein LOC105649755 [Jatropha curcas] |
| 14 |
Hb_003216_090 |
0.0678928639 |
- |
- |
PREDICTED: nuclear pore complex protein NUP133 isoform X1 [Jatropha curcas] |
| 15 |
Hb_001341_160 |
0.067943161 |
- |
- |
PREDICTED: E3 SUMO-protein ligase SIZ1 isoform X2 [Jatropha curcas] |
| 16 |
Hb_000066_150 |
0.0682218965 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein POPTR_0004s20690g [Populus trichocarpa] |
| 17 |
Hb_001673_050 |
0.0699374351 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 8-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_000331_580 |
0.0702860193 |
- |
- |
PREDICTED: programmed cell death protein 4-like [Jatropha curcas] |
| 19 |
Hb_011202_030 |
0.0708331637 |
- |
- |
arsenite-resistance protein, putative [Ricinus communis] |
| 20 |
Hb_123531_010 |
0.0720824251 |
- |
- |
PREDICTED: nuclear pore complex protein NUP98A isoform X1 [Jatropha curcas] |