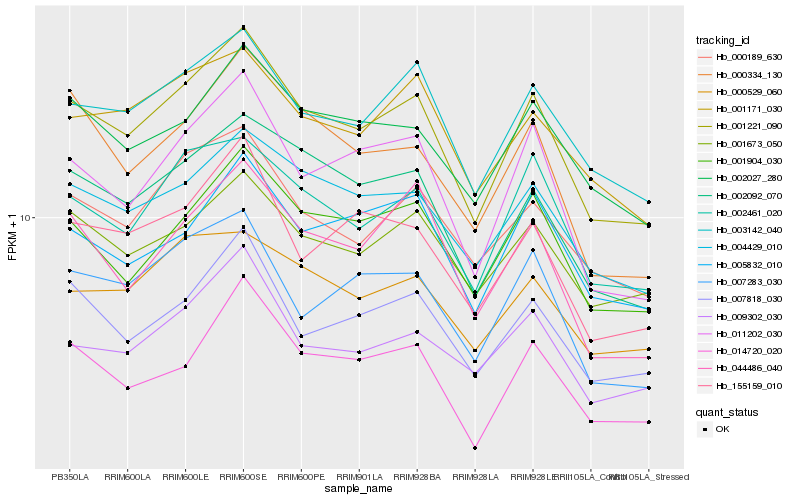
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001221_090 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105640211 isoform X1 [Jatropha curcas] |
| 2 |
Hb_003142_040 |
0.0550578667 |
- |
- |
Sf3a3 [Gossypium arboreum] |
| 3 |
Hb_002092_070 |
0.0570834864 |
- |
- |
PREDICTED: aluminum-activated malate transporter 9-like [Jatropha curcas] |
| 4 |
Hb_004429_010 |
0.0585040078 |
- |
- |
calcineurin-like phosphoesterase [Manihot esculenta] |
| 5 |
Hb_011202_030 |
0.061275009 |
- |
- |
arsenite-resistance protein, putative [Ricinus communis] |
| 6 |
Hb_001904_030 |
0.0612886608 |
- |
- |
PREDICTED: importin-5 [Jatropha curcas] |
| 7 |
Hb_002461_020 |
0.0620607937 |
- |
- |
PREDICTED: uncharacterized protein LOC105642649 isoform X2 [Jatropha curcas] |
| 8 |
Hb_007818_030 |
0.0637640636 |
- |
- |
PREDICTED: transcriptional corepressor SEUSS-like [Jatropha curcas] |
| 9 |
Hb_002027_280 |
0.064739971 |
- |
- |
PREDICTED: sister-chromatid cohesion protein 3 [Jatropha curcas] |
| 10 |
Hb_000189_630 |
0.0648386472 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1 [Jatropha curcas] |
| 11 |
Hb_001673_050 |
0.0684104136 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 8-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_014720_020 |
0.0685770542 |
- |
- |
PREDICTED: serine/threonine-protein kinase ATM [Jatropha curcas] |
| 13 |
Hb_155159_010 |
0.0698034568 |
- |
- |
PREDICTED: RNA polymerase II C-terminal domain phosphatase-like 1 [Jatropha curcas] |
| 14 |
Hb_001171_030 |
0.0702516821 |
transcription factor |
TF Family: LUG |
PREDICTED: transcriptional corepressor LEUNIG-like [Jatropha curcas] |
| 15 |
Hb_007283_030 |
0.0709190325 |
- |
- |
PREDICTED: pre-mRNA-processing protein 40C-like [Malus domestica] |
| 16 |
Hb_000334_130 |
0.0728656464 |
- |
- |
hypothetical protein JCGZ_16781 [Jatropha curcas] |
| 17 |
Hb_005832_010 |
0.073049534 |
- |
- |
PREDICTED: enhancer of mRNA-decapping protein 4 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000529_060 |
0.0732580942 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 17 isoform X1 [Jatropha curcas] |
| 19 |
Hb_009302_030 |
0.0733489148 |
- |
- |
hypothetical protein RCOM_1176340 [Ricinus communis] |
| 20 |
Hb_044486_040 |
0.0734902469 |
- |
- |
PREDICTED: ion channel CASTOR-like [Malus domestica] |