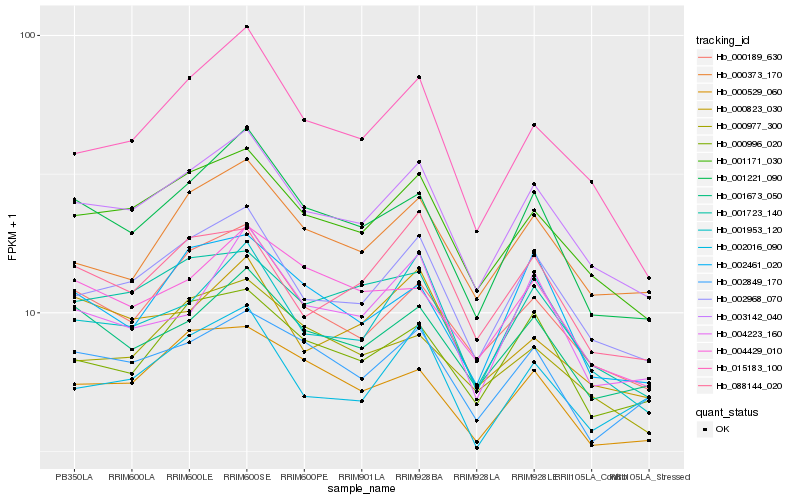
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003142_040 |
0.0 |
- |
- |
Sf3a3 [Gossypium arboreum] |
| 2 |
Hb_002968_070 |
0.0305574894 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 4b isoform X2 [Jatropha curcas] |
| 3 |
Hb_001171_030 |
0.0323759933 |
transcription factor |
TF Family: LUG |
PREDICTED: transcriptional corepressor LEUNIG-like [Jatropha curcas] |
| 4 |
Hb_001953_120 |
0.0539545312 |
- |
- |
PREDICTED: uncharacterized protein LOC105648761 isoform X1 [Jatropha curcas] |
| 5 |
Hb_001221_090 |
0.0550578667 |
- |
- |
PREDICTED: uncharacterized protein LOC105640211 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000189_630 |
0.0555682699 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1 [Jatropha curcas] |
| 7 |
Hb_001723_140 |
0.059541086 |
- |
- |
PREDICTED: importin beta-like SAD2 homolog isoform X2 [Jatropha curcas] |
| 8 |
Hb_001673_050 |
0.0600048127 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 8-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_000373_170 |
0.0640554351 |
- |
- |
PREDICTED: exocyst complex component SEC5A-like [Jatropha curcas] |
| 10 |
Hb_000996_020 |
0.0643795218 |
- |
- |
PREDICTED: RNA-binding protein NOB1 [Jatropha curcas] |
| 11 |
Hb_004223_160 |
0.0677407922 |
- |
- |
PREDICTED: 5'-3' exoribonuclease 3 isoform X2 [Jatropha curcas] |
| 12 |
Hb_000529_060 |
0.0680648628 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 17 isoform X1 [Jatropha curcas] |
| 13 |
Hb_088144_020 |
0.0688336878 |
transcription factor |
TF Family: ARID |
PREDICTED: AT-rich interactive domain-containing protein 6-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_002016_090 |
0.0695520967 |
- |
- |
PREDICTED: DNA mismatch repair protein PMS1 [Jatropha curcas] |
| 15 |
Hb_015183_100 |
0.0696706815 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At3g12360 [Jatropha curcas] |
| 16 |
Hb_000977_300 |
0.0708657124 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: probable E3 ubiquitin-protein ligase ARI8 [Malus domestica] |
| 17 |
Hb_000823_030 |
0.0713459108 |
- |
- |
PREDICTED: DNA-directed RNA polymerase III subunit RPC5 [Jatropha curcas] |
| 18 |
Hb_002849_170 |
0.0714318223 |
- |
- |
PREDICTED: uncharacterized protein LOC105642955 isoform X1 [Jatropha curcas] |
| 19 |
Hb_002461_020 |
0.0714563888 |
- |
- |
PREDICTED: uncharacterized protein LOC105642649 isoform X2 [Jatropha curcas] |
| 20 |
Hb_004429_010 |
0.0724971412 |
- |
- |
calcineurin-like phosphoesterase [Manihot esculenta] |