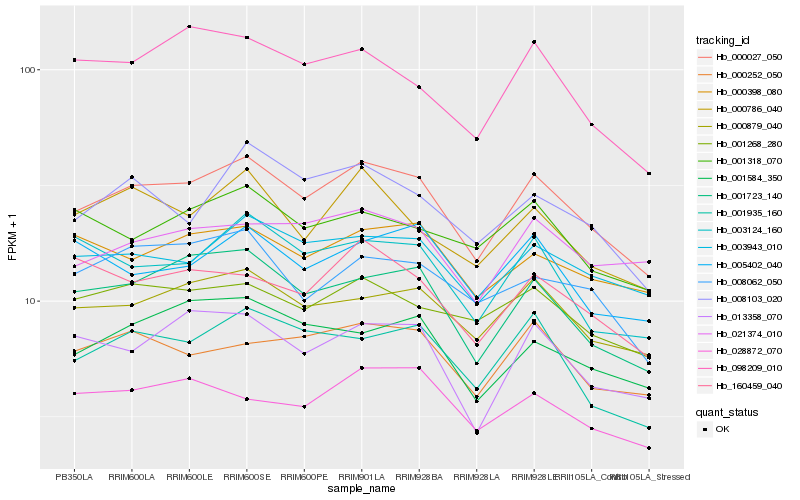
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000027_050 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105636933 isoform X1 [Jatropha curcas] |
| 2 |
Hb_000879_040 |
0.061374309 |
- |
- |
PREDICTED: calcium-transporting ATPase 3, endoplasmic reticulum-type [Jatropha curcas] |
| 3 |
Hb_001723_140 |
0.0624918338 |
- |
- |
PREDICTED: importin beta-like SAD2 homolog isoform X2 [Jatropha curcas] |
| 4 |
Hb_003943_010 |
0.0654659108 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_001935_160 |
0.0727902397 |
- |
- |
PREDICTED: F-box protein At5g51370-like [Jatropha curcas] |
| 6 |
Hb_000252_050 |
0.0731858837 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g60770 [Jatropha curcas] |
| 7 |
Hb_001268_280 |
0.0751120976 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_013358_070 |
0.0755279524 |
- |
- |
protein with unknown function [Ricinus communis] |
| 9 |
Hb_000786_040 |
0.0763250113 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 17 [Jatropha curcas] |
| 10 |
Hb_008103_020 |
0.0770811416 |
- |
- |
hypothetical protein POPTR_0003s20310g [Populus trichocarpa] |
| 11 |
Hb_001584_350 |
0.0778839113 |
- |
- |
hypothetical protein JCGZ_23178 [Jatropha curcas] |
| 12 |
Hb_021374_010 |
0.0786499263 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC transcription factor NAM-A1 [Populus euphratica] |
| 13 |
Hb_000398_080 |
0.0799099315 |
- |
- |
tip120, putative [Ricinus communis] |
| 14 |
Hb_008062_050 |
0.0807255224 |
- |
- |
PREDICTED: uncharacterized protein LOC105631631 [Jatropha curcas] |
| 15 |
Hb_003124_160 |
0.0810331041 |
- |
- |
dynamin, putative [Ricinus communis] |
| 16 |
Hb_028872_070 |
0.0815326305 |
desease resistance |
Gene Name: DEAD |
PREDICTED: putative pre-mRNA-splicing factor ATP-dependent RNA helicase PRP1 isoform X1 [Jatropha curcas] |
| 17 |
Hb_098209_010 |
0.0821317414 |
- |
- |
ATP synthase subunit beta vacuolar, putative [Ricinus communis] |
| 18 |
Hb_160459_040 |
0.0822237649 |
- |
- |
hypothetical protein JCGZ_01511 [Jatropha curcas] |
| 19 |
Hb_001318_070 |
0.0839853166 |
transcription factor |
TF Family: SBP |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_005402_040 |
0.0842538571 |
- |
- |
PREDICTED: replication factor C subunit 1 [Jatropha curcas] |