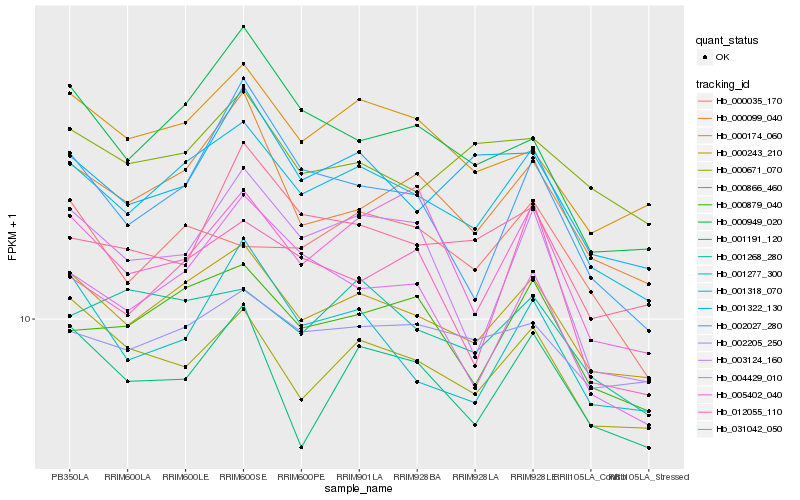
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001318_070 |
0.0 |
transcription factor |
TF Family: SBP |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000243_210 |
0.0282058146 |
- |
- |
protein with unknown function [Ricinus communis] |
| 3 |
Hb_000879_040 |
0.0500191801 |
- |
- |
PREDICTED: calcium-transporting ATPase 3, endoplasmic reticulum-type [Jatropha curcas] |
| 4 |
Hb_000099_040 |
0.0571178529 |
- |
- |
unnamed protein product [Coffea canephora] |
| 5 |
Hb_000174_060 |
0.0625387197 |
- |
- |
zinc finger family protein [Populus trichocarpa] |
| 6 |
Hb_001268_280 |
0.0641855502 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_002205_250 |
0.0646993141 |
- |
- |
PREDICTED: probable inactive serine/threonine-protein kinase scy1 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000671_070 |
0.0655802503 |
- |
- |
PREDICTED: uncharacterized protein LOC105630621 [Jatropha curcas] |
| 9 |
Hb_001322_130 |
0.06583177 |
- |
- |
PREDICTED: putative glycerol-3-phosphate transporter 4 [Jatropha curcas] |
| 10 |
Hb_001277_300 |
0.0692670312 |
- |
- |
PREDICTED: uncharacterized protein LOC105632051 isoform X4 [Jatropha curcas] |
| 11 |
Hb_000949_020 |
0.0703927884 |
transcription factor |
TF Family: Orphans |
PREDICTED: ethylene receptor isoform X2 [Jatropha curcas] |
| 12 |
Hb_005402_040 |
0.0709918582 |
- |
- |
PREDICTED: replication factor C subunit 1 [Jatropha curcas] |
| 13 |
Hb_000035_170 |
0.0713213199 |
- |
- |
PREDICTED: DNA-directed RNA polymerases IV and V subunit 2-like [Jatropha curcas] |
| 14 |
Hb_012055_110 |
0.0718298386 |
- |
- |
tetratricopeptide repeat protein, tpr, putative [Ricinus communis] |
| 15 |
Hb_001191_120 |
0.0720022774 |
- |
- |
PREDICTED: cleavage stimulating factor 64 isoform X1 [Jatropha curcas] |
| 16 |
Hb_003124_160 |
0.0721013554 |
- |
- |
dynamin, putative [Ricinus communis] |
| 17 |
Hb_031042_050 |
0.0723894426 |
- |
- |
PREDICTED: protein transport protein SEC16B homolog [Jatropha curcas] |
| 18 |
Hb_000866_460 |
0.0728699766 |
- |
- |
PREDICTED: acetyl-coenzyme A carboxylase carboxyl transferase subunit alpha, chloroplastic-like [Jatropha curcas] |
| 19 |
Hb_004429_010 |
0.0732537773 |
- |
- |
calcineurin-like phosphoesterase [Manihot esculenta] |
| 20 |
Hb_002027_280 |
0.0738972639 |
- |
- |
PREDICTED: sister-chromatid cohesion protein 3 [Jatropha curcas] |