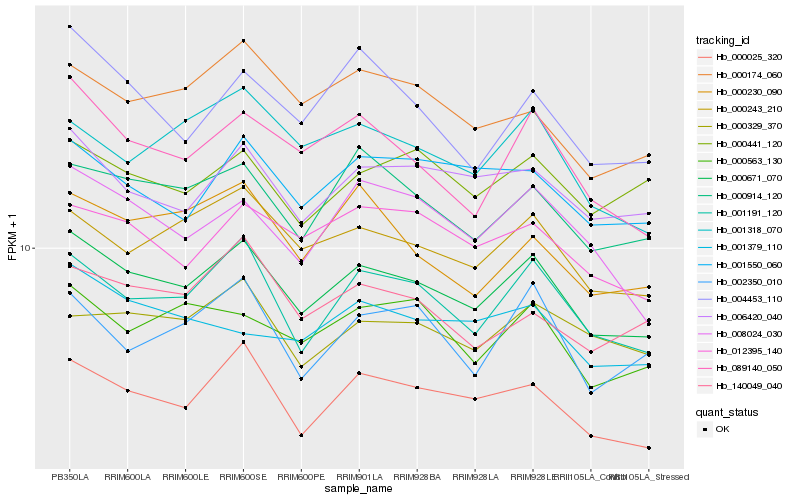
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000671_070 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105630621 [Jatropha curcas] |
| 2 |
Hb_001191_120 |
0.0447954388 |
- |
- |
PREDICTED: cleavage stimulating factor 64 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000025_320 |
0.0478595074 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g28010 [Jatropha curcas] |
| 4 |
Hb_000243_210 |
0.052478951 |
- |
- |
protein with unknown function [Ricinus communis] |
| 5 |
Hb_000914_120 |
0.0543482351 |
- |
- |
PREDICTED: outer envelope protein 80, chloroplastic [Jatropha curcas] |
| 6 |
Hb_089140_050 |
0.0571231915 |
- |
- |
hypothetical protein JCGZ_07485 [Jatropha curcas] |
| 7 |
Hb_006420_040 |
0.058309347 |
transcription factor |
TF Family: Orphans |
PREDICTED: paired amphipathic helix protein Sin3-like 4 isoform X2 [Jatropha curcas] |
| 8 |
Hb_012395_140 |
0.0615154353 |
- |
- |
PREDICTED: uncharacterized protein LOC105638411 [Jatropha curcas] |
| 9 |
Hb_000174_060 |
0.0637136156 |
- |
- |
zinc finger family protein [Populus trichocarpa] |
| 10 |
Hb_000563_130 |
0.063732272 |
- |
- |
PREDICTED: WD repeat-containing protein 43 [Jatropha curcas] |
| 11 |
Hb_008024_030 |
0.0642954061 |
transcription factor |
TF Family: Trihelix |
PREDICTED: uncharacterized protein LOC105648348 [Jatropha curcas] |
| 12 |
Hb_001550_060 |
0.065178177 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase BSL3 isoform X4 [Jatropha curcas] |
| 13 |
Hb_001318_070 |
0.0655802503 |
transcription factor |
TF Family: SBP |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_004453_110 |
0.0686984961 |
- |
- |
Peptidyl-prolyl cis-trans isomerase CYP19-2 isoform 1 [Theobroma cacao] |
| 15 |
Hb_140049_040 |
0.0689263385 |
- |
- |
PREDICTED: uncharacterized protein LOC105632012 [Jatropha curcas] |
| 16 |
Hb_000230_090 |
0.069393117 |
- |
- |
PREDICTED: WD repeat-containing protein 48 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000329_370 |
0.0696343979 |
- |
- |
hypothetical protein JCGZ_14571 [Jatropha curcas] |
| 18 |
Hb_002350_010 |
0.0699514047 |
- |
- |
PREDICTED: DNA-directed RNA polymerase III subunit rpc1 [Jatropha curcas] |
| 19 |
Hb_000441_120 |
0.070662398 |
- |
- |
PREDICTED: DNA-damage-repair/toleration protein DRT111, chloroplastic isoform X1 [Jatropha curcas] |
| 20 |
Hb_001379_110 |
0.0710051064 |
- |
- |
PREDICTED: sec1 family domain-containing protein MIP3 [Jatropha curcas] |