| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
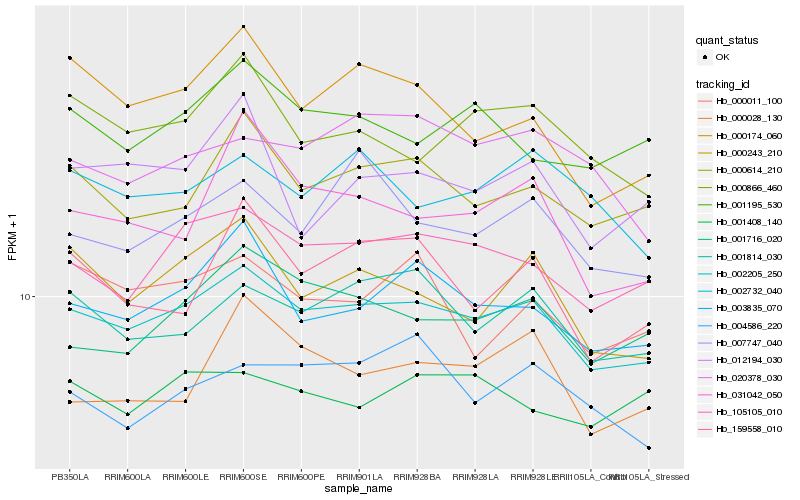
Hb_002205_250 |
0.0 |
- |
- |
PREDICTED: probable inactive serine/threonine-protein kinase scy1 isoform X1 [Jatropha curcas] |
| 2 |
Hb_105105_010 |
0.0404822665 |
- |
- |
PREDICTED: coiled-coil domain-containing protein 132 [Jatropha curcas] |
| 3 |
Hb_003835_070 |
0.0471879431 |
- |
- |
PREDICTED: uncharacterized protein LOC105641698 isoform X2 [Jatropha curcas] |
| 4 |
Hb_000614_210 |
0.0473248891 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor GT-1-like isoform X1 [Jatropha curcas] |
| 5 |
Hb_001814_030 |
0.0491245134 |
- |
- |
PREDICTED: uncharacterized protein LOC105650634 [Jatropha curcas] |
| 6 |
Hb_001716_020 |
0.0520174098 |
- |
- |
hydrolase, putative [Ricinus communis] |
| 7 |
Hb_000243_210 |
0.0525886065 |
- |
- |
protein with unknown function [Ricinus communis] |
| 8 |
Hb_000866_460 |
0.0550337989 |
- |
- |
PREDICTED: acetyl-coenzyme A carboxylase carboxyl transferase subunit alpha, chloroplastic-like [Jatropha curcas] |
| 9 |
Hb_001195_530 |
0.0566639281 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_002732_040 |
0.0573498556 |
- |
- |
PREDICTED: uncharacterized protein LOC105628163 isoform X1 [Jatropha curcas] |
| 11 |
Hb_007747_040 |
0.0581713224 |
- |
- |
PREDICTED: probable UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase SPINDLY [Jatropha curcas] |
| 12 |
Hb_020378_030 |
0.0606233794 |
- |
- |
PREDICTED: protein transport protein SEC31 homolog B [Jatropha curcas] |
| 13 |
Hb_004586_220 |
0.0612450173 |
- |
- |
PREDICTED: WD and tetratricopeptide repeats protein 1 isoform X3 [Jatropha curcas] |
| 14 |
Hb_001408_140 |
0.0612868501 |
- |
- |
hypothetical protein JCGZ_00234 [Jatropha curcas] |
| 15 |
Hb_000174_060 |
0.061327576 |
- |
- |
zinc finger family protein [Populus trichocarpa] |
| 16 |
Hb_000011_100 |
0.0614915566 |
- |
- |
PREDICTED: uncharacterized protein C630.12 [Jatropha curcas] |
| 17 |
Hb_012194_030 |
0.0618543925 |
- |
- |
PREDICTED: R3H domain-containing protein 2-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_159558_010 |
0.0623479339 |
- |
- |
PREDICTED: uncharacterized protein LOC105635846 [Jatropha curcas] |
| 19 |
Hb_000028_130 |
0.0628302264 |
- |
- |
PREDICTED: uncharacterized protein LOC105645303 isoform X1 [Jatropha curcas] |
| 20 |
Hb_031042_050 |
0.0628862847 |
- |
- |
PREDICTED: protein transport protein SEC16B homolog [Jatropha curcas] |