| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
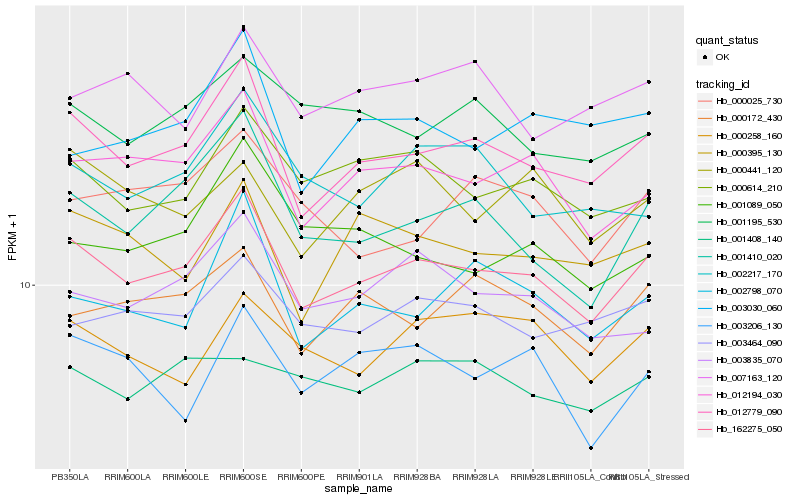
Hb_162275_050 |
0.0 |
- |
- |
PREDICTED: mRNA-capping enzyme [Jatropha curcas] |
| 2 |
Hb_012779_090 |
0.0300102568 |
- |
- |
PREDICTED: lipase member N [Jatropha curcas] |
| 3 |
Hb_012194_030 |
0.0475172942 |
- |
- |
PREDICTED: R3H domain-containing protein 2-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_000614_210 |
0.0562964752 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor GT-1-like isoform X1 [Jatropha curcas] |
| 5 |
Hb_001410_020 |
0.0565147601 |
- |
- |
cdk8, putative [Ricinus communis] |
| 6 |
Hb_003835_070 |
0.0595302748 |
- |
- |
PREDICTED: uncharacterized protein LOC105641698 isoform X2 [Jatropha curcas] |
| 7 |
Hb_003464_090 |
0.0609355576 |
transcription factor |
TF Family: Trihelix |
PREDICTED: uncharacterized protein LOC105642342 [Jatropha curcas] |
| 8 |
Hb_000172_430 |
0.0616288167 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_002798_070 |
0.0623319674 |
transcription factor |
TF Family: Jumonji |
PREDICTED: putative lysine-specific demethylase JMJ16 [Jatropha curcas] |
| 10 |
Hb_000395_130 |
0.0647164373 |
- |
- |
PREDICTED: N6-adenosine-methyltransferase MT-A70-like [Jatropha curcas] |
| 11 |
Hb_001195_530 |
0.0656542903 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000258_160 |
0.0664848792 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_002217_170 |
0.0680984336 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase UPL6-like isoform X1 [Gossypium raimondii] |
| 14 |
Hb_003206_130 |
0.0685357253 |
- |
- |
PREDICTED: protein NRDE2 homolog [Jatropha curcas] |
| 15 |
Hb_001089_050 |
0.0685423003 |
transcription factor |
TF Family: ARID |
PREDICTED: AT-rich interactive domain-containing protein 4 [Jatropha curcas] |
| 16 |
Hb_000025_730 |
0.0687068514 |
- |
- |
PREDICTED: 60S ribosomal protein L2, mitochondrial [Jatropha curcas] |
| 17 |
Hb_000441_120 |
0.0687069083 |
- |
- |
PREDICTED: DNA-damage-repair/toleration protein DRT111, chloroplastic isoform X1 [Jatropha curcas] |
| 18 |
Hb_003030_060 |
0.069606817 |
- |
- |
PREDICTED: regulation of nuclear pre-mRNA domain-containing protein 2-like [Jatropha curcas] |
| 19 |
Hb_001408_140 |
0.0712841763 |
- |
- |
hypothetical protein JCGZ_00234 [Jatropha curcas] |
| 20 |
Hb_007163_120 |
0.0717235187 |
- |
- |
PREDICTED: COMPASS-like H3K4 histone methylase component WDR5A [Jatropha curcas] |