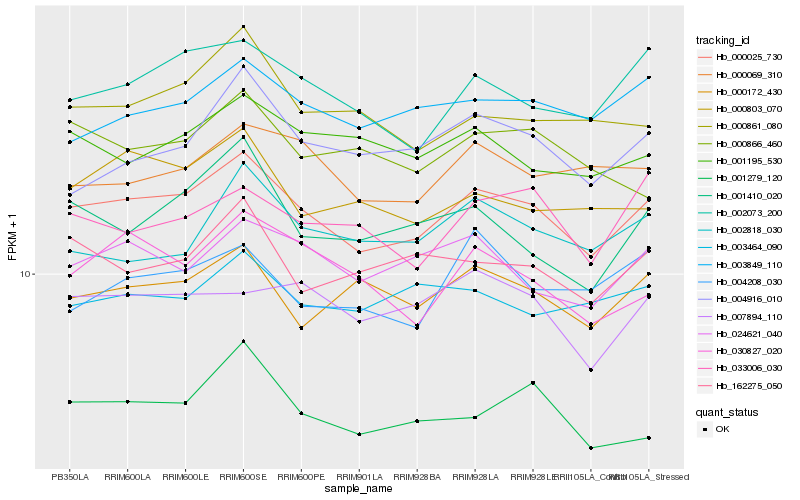
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000025_730 |
0.0 |
- |
- |
PREDICTED: 60S ribosomal protein L2, mitochondrial [Jatropha curcas] |
| 2 |
Hb_002073_200 |
0.0607868969 |
- |
- |
Phosphatase 2A-2 [Theobroma cacao] |
| 3 |
Hb_024621_040 |
0.0641402417 |
transcription factor |
TF Family: ERF |
hypothetical protein L484_014666 [Morus notabilis] |
| 4 |
Hb_003849_110 |
0.0660162112 |
- |
- |
hypothetical protein JCGZ_17842 [Jatropha curcas] |
| 5 |
Hb_000172_430 |
0.0670063242 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000069_310 |
0.0677997325 |
- |
- |
ubiquitin ligase [Cucumis melo subsp. melo] |
| 7 |
Hb_162275_050 |
0.0687068514 |
- |
- |
PREDICTED: mRNA-capping enzyme [Jatropha curcas] |
| 8 |
Hb_001195_530 |
0.0695132566 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000803_070 |
0.0703019939 |
- |
- |
PREDICTED: protein TRAUCO [Jatropha curcas] |
| 10 |
Hb_030827_020 |
0.0704134304 |
- |
- |
PREDICTED: protein ROOT PRIMORDIUM DEFECTIVE 1 [Jatropha curcas] |
| 11 |
Hb_007894_110 |
0.0705797378 |
- |
- |
Heat-shock protein 105 kDa, putative [Ricinus communis] |
| 12 |
Hb_003464_090 |
0.07088953 |
transcription factor |
TF Family: Trihelix |
PREDICTED: uncharacterized protein LOC105642342 [Jatropha curcas] |
| 13 |
Hb_000861_080 |
0.0710306857 |
- |
- |
PREDICTED: protein ELC-like [Jatropha curcas] |
| 14 |
Hb_001279_120 |
0.0710349547 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 5-like [Jatropha curcas] |
| 15 |
Hb_001410_020 |
0.0710915454 |
- |
- |
cdk8, putative [Ricinus communis] |
| 16 |
Hb_004916_010 |
0.0720601682 |
- |
- |
PREDICTED: uncharacterized protein LOC105631086 [Jatropha curcas] |
| 17 |
Hb_033006_030 |
0.0730726219 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At1g22040-like [Jatropha curcas] |
| 18 |
Hb_002818_030 |
0.0741770238 |
- |
- |
PREDICTED: protein transport protein SEC23 [Jatropha curcas] |
| 19 |
Hb_000866_460 |
0.0773207862 |
- |
- |
PREDICTED: acetyl-coenzyme A carboxylase carboxyl transferase subunit alpha, chloroplastic-like [Jatropha curcas] |
| 20 |
Hb_004208_030 |
0.0773954266 |
transcription factor |
TF Family: MYB-related |
PREDICTED: TSL-kinase interacting protein 1 [Jatropha curcas] |