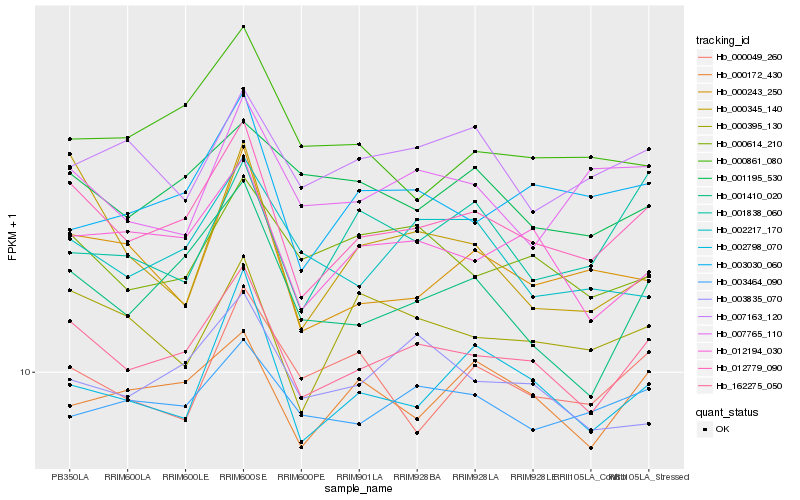
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012779_090 |
0.0 |
- |
- |
PREDICTED: lipase member N [Jatropha curcas] |
| 2 |
Hb_162275_050 |
0.0300102568 |
- |
- |
PREDICTED: mRNA-capping enzyme [Jatropha curcas] |
| 3 |
Hb_002798_070 |
0.0554111641 |
transcription factor |
TF Family: Jumonji |
PREDICTED: putative lysine-specific demethylase JMJ16 [Jatropha curcas] |
| 4 |
Hb_000243_250 |
0.0594149133 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 44 [Jatropha curcas] |
| 5 |
Hb_012194_030 |
0.0601586213 |
- |
- |
PREDICTED: R3H domain-containing protein 2-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_000395_130 |
0.0615376102 |
- |
- |
PREDICTED: N6-adenosine-methyltransferase MT-A70-like [Jatropha curcas] |
| 7 |
Hb_001410_020 |
0.0623189519 |
- |
- |
cdk8, putative [Ricinus communis] |
| 8 |
Hb_000172_430 |
0.0631507063 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_003030_060 |
0.0675977231 |
- |
- |
PREDICTED: regulation of nuclear pre-mRNA domain-containing protein 2-like [Jatropha curcas] |
| 10 |
Hb_003464_090 |
0.0689975569 |
transcription factor |
TF Family: Trihelix |
PREDICTED: uncharacterized protein LOC105642342 [Jatropha curcas] |
| 11 |
Hb_007765_110 |
0.0715220946 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 40-like [Jatropha curcas] |
| 12 |
Hb_007163_120 |
0.0731189292 |
- |
- |
PREDICTED: COMPASS-like H3K4 histone methylase component WDR5A [Jatropha curcas] |
| 13 |
Hb_000614_210 |
0.0741958006 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor GT-1-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_001195_530 |
0.0748547139 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_002217_170 |
0.0754966097 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase UPL6-like isoform X1 [Gossypium raimondii] |
| 16 |
Hb_003835_070 |
0.0755645328 |
- |
- |
PREDICTED: uncharacterized protein LOC105641698 isoform X2 [Jatropha curcas] |
| 17 |
Hb_000861_080 |
0.0761476182 |
- |
- |
PREDICTED: protein ELC-like [Jatropha curcas] |
| 18 |
Hb_000049_260 |
0.0761914111 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 19 |
Hb_000345_140 |
0.0763214146 |
- |
- |
PREDICTED: autophagy-related protein 18a-like [Jatropha curcas] |
| 20 |
Hb_001838_060 |
0.076462378 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor HBP-1b(c38)-like isoform X3 [Jatropha curcas] |