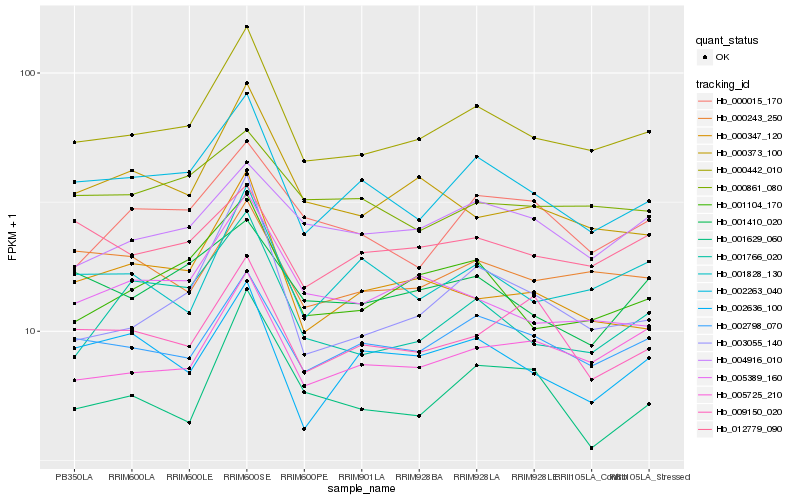
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000442_010 |
0.0 |
- |
- |
hypothetical protein POPTR_0010s19060g [Populus trichocarpa] |
| 2 |
Hb_002798_070 |
0.0623885668 |
transcription factor |
TF Family: Jumonji |
PREDICTED: putative lysine-specific demethylase JMJ16 [Jatropha curcas] |
| 3 |
Hb_002263_040 |
0.0627747713 |
- |
- |
carbon catabolite repressor protein, putative [Ricinus communis] |
| 4 |
Hb_001104_170 |
0.0629663232 |
transcription factor |
TF Family: HSF |
PREDICTED: heat stress transcription factor A-2c [Jatropha curcas] |
| 5 |
Hb_005725_210 |
0.0660552649 |
- |
- |
PREDICTED: RNA-binding protein 1 [Jatropha curcas] |
| 6 |
Hb_005389_160 |
0.0764445572 |
- |
- |
PREDICTED: casein kinase I [Jatropha curcas] |
| 7 |
Hb_000243_250 |
0.0772003237 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 44 [Jatropha curcas] |
| 8 |
Hb_003055_140 |
0.078292104 |
transcription factor |
TF Family: SNF2 |
PREDICTED: uncharacterized protein LOC105649495 [Jatropha curcas] |
| 9 |
Hb_001828_130 |
0.0793658479 |
- |
- |
PREDICTED: transcription factor GTE8-like [Jatropha curcas] |
| 10 |
Hb_001766_020 |
0.0798308399 |
- |
- |
PREDICTED: probable galacturonosyltransferase 14 [Populus euphratica] |
| 11 |
Hb_004916_010 |
0.0835515 |
- |
- |
PREDICTED: uncharacterized protein LOC105631086 [Jatropha curcas] |
| 12 |
Hb_012779_090 |
0.0852483629 |
- |
- |
PREDICTED: lipase member N [Jatropha curcas] |
| 13 |
Hb_000015_170 |
0.0853645116 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_001410_020 |
0.0854006398 |
- |
- |
cdk8, putative [Ricinus communis] |
| 15 |
Hb_000347_120 |
0.0860353419 |
- |
- |
PREDICTED: uncharacterized protein LOC105631363 [Jatropha curcas] |
| 16 |
Hb_000861_080 |
0.0875513127 |
- |
- |
PREDICTED: protein ELC-like [Jatropha curcas] |
| 17 |
Hb_000373_100 |
0.0880736072 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor VIP1 [Jatropha curcas] |
| 18 |
Hb_001629_060 |
0.0881063416 |
transcription factor |
TF Family: SWI/SNF-SWI3 |
Flavin-containing amine oxidase domain-containing protein, putative [Ricinus communis] |
| 19 |
Hb_009150_020 |
0.0881492349 |
- |
- |
PREDICTED: uncharacterized protein LOC105630583 [Jatropha curcas] |
| 20 |
Hb_002636_100 |
0.09095635 |
- |
- |
PREDICTED: DNA-directed RNA polymerase 1B, mitochondrial [Jatropha curcas] |