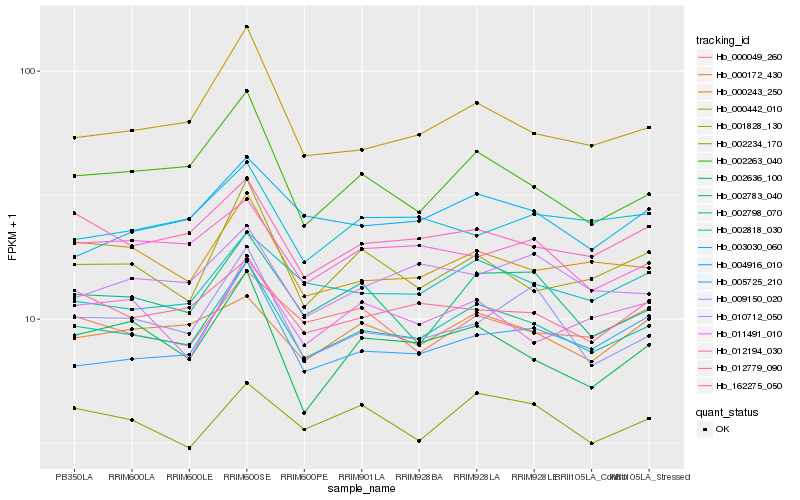
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002798_070 |
0.0 |
transcription factor |
TF Family: Jumonji |
PREDICTED: putative lysine-specific demethylase JMJ16 [Jatropha curcas] |
| 2 |
Hb_012779_090 |
0.0554111641 |
- |
- |
PREDICTED: lipase member N [Jatropha curcas] |
| 3 |
Hb_000243_250 |
0.0579273057 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 44 [Jatropha curcas] |
| 4 |
Hb_002818_030 |
0.0594178228 |
- |
- |
PREDICTED: protein transport protein SEC23 [Jatropha curcas] |
| 5 |
Hb_002783_040 |
0.062187227 |
transcription factor |
TF Family: IWS1 |
- |
| 6 |
Hb_162275_050 |
0.0623319674 |
- |
- |
PREDICTED: mRNA-capping enzyme [Jatropha curcas] |
| 7 |
Hb_000442_010 |
0.0623885668 |
- |
- |
hypothetical protein POPTR_0010s19060g [Populus trichocarpa] |
| 8 |
Hb_005725_210 |
0.065051727 |
- |
- |
PREDICTED: RNA-binding protein 1 [Jatropha curcas] |
| 9 |
Hb_002263_040 |
0.0652525503 |
- |
- |
carbon catabolite repressor protein, putative [Ricinus communis] |
| 10 |
Hb_000172_430 |
0.0655499902 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_002234_170 |
0.0656364762 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 5-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_001828_130 |
0.0658441306 |
- |
- |
PREDICTED: transcription factor GTE8-like [Jatropha curcas] |
| 13 |
Hb_000049_260 |
0.0675693509 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 14 |
Hb_004916_010 |
0.068181336 |
- |
- |
PREDICTED: uncharacterized protein LOC105631086 [Jatropha curcas] |
| 15 |
Hb_009150_020 |
0.0696771722 |
- |
- |
PREDICTED: uncharacterized protein LOC105630583 [Jatropha curcas] |
| 16 |
Hb_012194_030 |
0.070460588 |
- |
- |
PREDICTED: R3H domain-containing protein 2-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_011491_010 |
0.0716743368 |
- |
- |
Protein YME1, putative [Ricinus communis] |
| 18 |
Hb_010712_050 |
0.0717131033 |
- |
- |
PREDICTED: protein ELC-like [Jatropha curcas] |
| 19 |
Hb_002636_100 |
0.0738343499 |
- |
- |
PREDICTED: DNA-directed RNA polymerase 1B, mitochondrial [Jatropha curcas] |
| 20 |
Hb_003030_060 |
0.074225664 |
- |
- |
PREDICTED: regulation of nuclear pre-mRNA domain-containing protein 2-like [Jatropha curcas] |