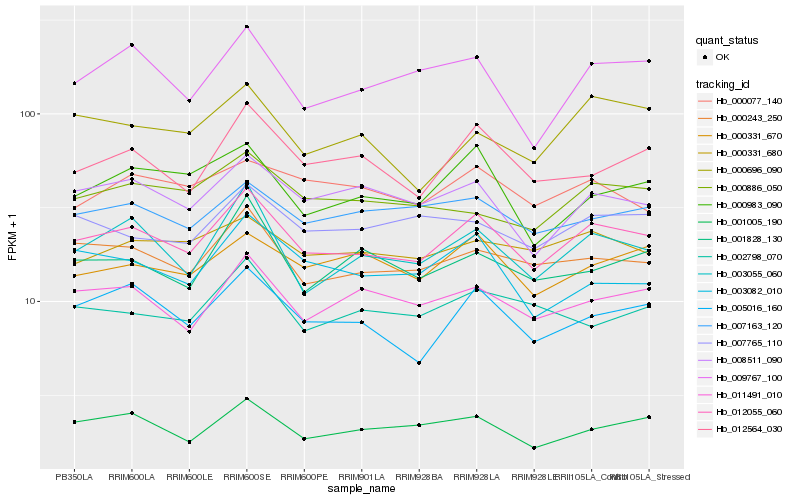
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012055_060 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105639799 isoform X1 [Jatropha curcas] |
| 2 |
Hb_000243_250 |
0.0658309481 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 44 [Jatropha curcas] |
| 3 |
Hb_005016_160 |
0.066697542 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 8 [Jatropha curcas] |
| 4 |
Hb_003055_060 |
0.0698039257 |
transcription factor |
TF Family: MYB-related |
PREDICTED: uncharacterized protein LOC105649487 isoform X1 [Jatropha curcas] |
| 5 |
Hb_001005_190 |
0.0727854508 |
- |
- |
PREDICTED: UPF0496 protein At3g19330-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_011491_010 |
0.0729398079 |
- |
- |
Protein YME1, putative [Ricinus communis] |
| 7 |
Hb_008511_090 |
0.0729409845 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At3g05675-like [Jatropha curcas] |
| 8 |
Hb_012564_030 |
0.0745789728 |
- |
- |
coronatine-insensitive 1 [Hevea brasiliensis] |
| 9 |
Hb_000331_670 |
0.0786341331 |
- |
- |
PREDICTED: protein tipD [Jatropha curcas] |
| 10 |
Hb_007163_120 |
0.0793787133 |
- |
- |
PREDICTED: COMPASS-like H3K4 histone methylase component WDR5A [Jatropha curcas] |
| 11 |
Hb_009767_100 |
0.0799746247 |
- |
- |
PREDICTED: FRIGIDA-like protein 4a [Jatropha curcas] |
| 12 |
Hb_000983_090 |
0.0810047743 |
- |
- |
PREDICTED: mitogen-activated protein kinase homolog NTF3 [Jatropha curcas] |
| 13 |
Hb_000331_680 |
0.0821263259 |
- |
- |
Protein bem46, putative [Ricinus communis] |
| 14 |
Hb_000886_050 |
0.0844697418 |
- |
- |
hypothetical protein POPTR_0003s01110g [Populus trichocarpa] |
| 15 |
Hb_007765_110 |
0.0850601341 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 40-like [Jatropha curcas] |
| 16 |
Hb_003082_010 |
0.0861116674 |
- |
- |
PREDICTED: putative F-box protein At3g10240 [Jatropha curcas] |
| 17 |
Hb_001828_130 |
0.0873303487 |
- |
- |
PREDICTED: transcription factor GTE8-like [Jatropha curcas] |
| 18 |
Hb_000077_140 |
0.0874792579 |
- |
- |
structural constituent of cell wall, putative [Ricinus communis] |
| 19 |
Hb_002798_070 |
0.0887905197 |
transcription factor |
TF Family: Jumonji |
PREDICTED: putative lysine-specific demethylase JMJ16 [Jatropha curcas] |
| 20 |
Hb_000696_090 |
0.0899921205 |
- |
- |
PREDICTED: uncharacterized protein LOC105647093 [Jatropha curcas] |