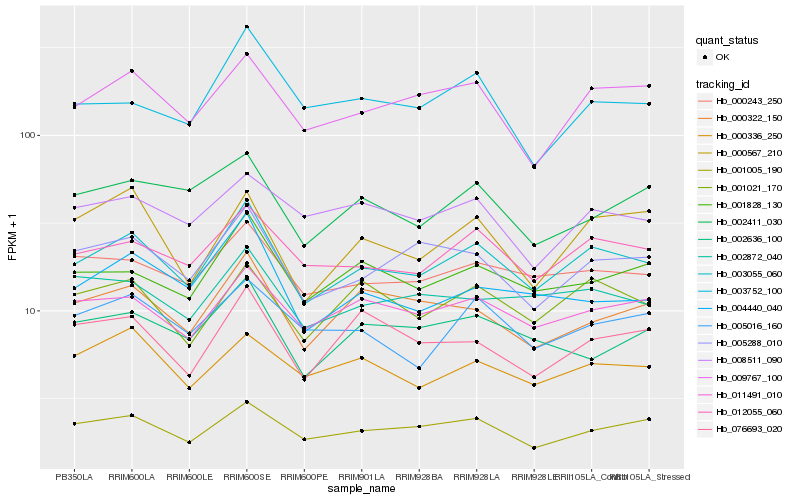
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003055_060 |
0.0 |
transcription factor |
TF Family: MYB-related |
PREDICTED: uncharacterized protein LOC105649487 isoform X1 [Jatropha curcas] |
| 2 |
Hb_012055_060 |
0.0698039257 |
- |
- |
PREDICTED: uncharacterized protein LOC105639799 isoform X1 [Jatropha curcas] |
| 3 |
Hb_009767_100 |
0.0757331913 |
- |
- |
PREDICTED: FRIGIDA-like protein 4a [Jatropha curcas] |
| 4 |
Hb_005288_010 |
0.0798256948 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 5 |
Hb_000243_250 |
0.0801558881 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 44 [Jatropha curcas] |
| 6 |
Hb_011491_010 |
0.0806963985 |
- |
- |
Protein YME1, putative [Ricinus communis] |
| 7 |
Hb_001005_190 |
0.081414902 |
- |
- |
PREDICTED: UPF0496 protein At3g19330-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_001828_130 |
0.0823269738 |
- |
- |
PREDICTED: transcription factor GTE8-like [Jatropha curcas] |
| 9 |
Hb_001021_170 |
0.0833832909 |
- |
- |
PREDICTED: uncharacterized protein LOC105642474 [Jatropha curcas] |
| 10 |
Hb_002872_040 |
0.0899497934 |
- |
- |
pumilio, putative [Ricinus communis] |
| 11 |
Hb_000322_150 |
0.0903374195 |
transcription factor |
TF Family: TRAF |
PREDICTED: regulatory protein NPR1 isoform X1 [Jatropha curcas] |
| 12 |
Hb_004440_040 |
0.0909104392 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 13 |
Hb_005016_160 |
0.0939345587 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 8 [Jatropha curcas] |
| 14 |
Hb_000336_250 |
0.0944523295 |
transcription factor |
TF Family: mTERF |
hypothetical protein B456_002G027700 [Gossypium raimondii] |
| 15 |
Hb_000567_210 |
0.0976571877 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor ASIL2-like [Jatropha curcas] |
| 16 |
Hb_002636_100 |
0.0978324195 |
- |
- |
PREDICTED: DNA-directed RNA polymerase 1B, mitochondrial [Jatropha curcas] |
| 17 |
Hb_003752_100 |
0.0985599724 |
- |
- |
PREDICTED: FRIGIDA-like protein 3 [Jatropha curcas] |
| 18 |
Hb_002411_030 |
0.0986789699 |
transcription factor |
TF Family: NF-YC |
PREDICTED: dr1-associated corepressor homolog [Jatropha curcas] |
| 19 |
Hb_076693_020 |
0.0991634022 |
- |
- |
hypothetical protein CISIN_1g0121812mg, partial [Citrus sinensis] |
| 20 |
Hb_008511_090 |
0.0997542966 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At3g05675-like [Jatropha curcas] |