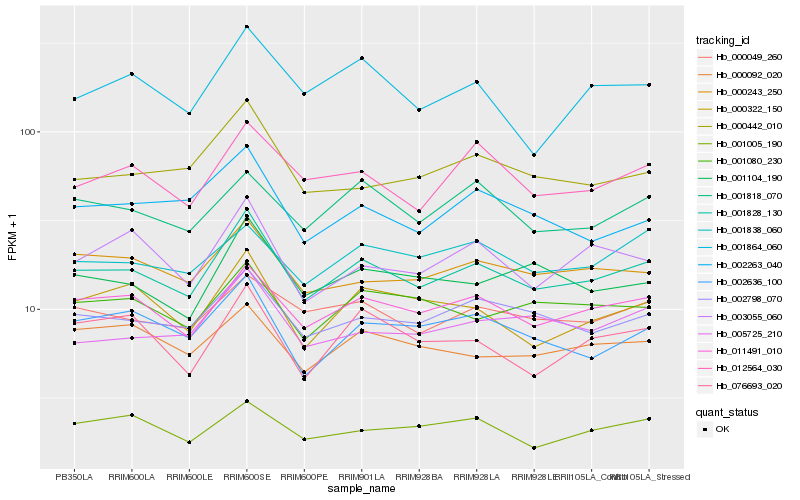
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001828_130 |
0.0 |
- |
- |
PREDICTED: transcription factor GTE8-like [Jatropha curcas] |
| 2 |
Hb_011491_010 |
0.0553036407 |
- |
- |
Protein YME1, putative [Ricinus communis] |
| 3 |
Hb_002798_070 |
0.0658441306 |
transcription factor |
TF Family: Jumonji |
PREDICTED: putative lysine-specific demethylase JMJ16 [Jatropha curcas] |
| 4 |
Hb_000243_250 |
0.0732940437 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 44 [Jatropha curcas] |
| 5 |
Hb_000322_150 |
0.0758952937 |
transcription factor |
TF Family: TRAF |
PREDICTED: regulatory protein NPR1 isoform X1 [Jatropha curcas] |
| 6 |
Hb_002636_100 |
0.0761594129 |
- |
- |
PREDICTED: DNA-directed RNA polymerase 1B, mitochondrial [Jatropha curcas] |
| 7 |
Hb_012564_030 |
0.0777639871 |
- |
- |
coronatine-insensitive 1 [Hevea brasiliensis] |
| 8 |
Hb_000049_260 |
0.0787419501 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 9 |
Hb_001104_190 |
0.0788814574 |
- |
- |
PREDICTED: uncharacterized protein LOC105629451 [Jatropha curcas] |
| 10 |
Hb_005725_210 |
0.0789891474 |
- |
- |
PREDICTED: RNA-binding protein 1 [Jatropha curcas] |
| 11 |
Hb_000442_010 |
0.0793658479 |
- |
- |
hypothetical protein POPTR_0010s19060g [Populus trichocarpa] |
| 12 |
Hb_002263_040 |
0.081704225 |
- |
- |
carbon catabolite repressor protein, putative [Ricinus communis] |
| 13 |
Hb_001818_070 |
0.0821247931 |
- |
- |
carbon catabolite repressor protein, putative [Ricinus communis] |
| 14 |
Hb_003055_060 |
0.0823269738 |
transcription factor |
TF Family: MYB-related |
PREDICTED: uncharacterized protein LOC105649487 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000092_020 |
0.0829425745 |
transcription factor |
TF Family: MYB |
transcription factor, putative [Ricinus communis] |
| 16 |
Hb_001080_230 |
0.0834142462 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105649979 [Jatropha curcas] |
| 17 |
Hb_001005_190 |
0.0848442573 |
- |
- |
PREDICTED: UPF0496 protein At3g19330-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_001864_060 |
0.085312491 |
transcription factor |
TF Family: GeBP |
PREDICTED: mediator-associated protein 1-like [Jatropha curcas] |
| 19 |
Hb_076693_020 |
0.0853314725 |
- |
- |
hypothetical protein CISIN_1g0121812mg, partial [Citrus sinensis] |
| 20 |
Hb_001838_060 |
0.0854573463 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor HBP-1b(c38)-like isoform X3 [Jatropha curcas] |