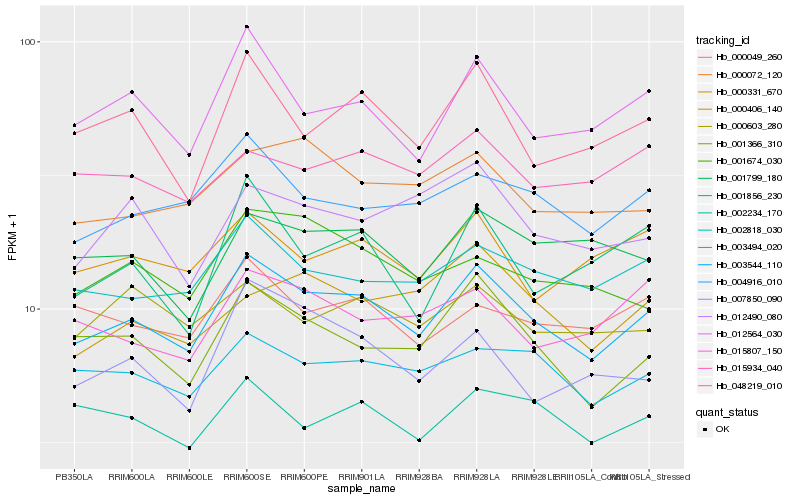
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003544_110 |
0.0 |
- |
- |
PREDICTED: zinc finger BED domain-containing protein RICESLEEPER 2-like isoform X1 [Jatropha curcas] |
| 2 |
Hb_001366_310 |
0.0692413196 |
- |
- |
PREDICTED: protein phosphatase 2C 32 [Jatropha curcas] |
| 3 |
Hb_012564_030 |
0.0719139243 |
- |
- |
coronatine-insensitive 1 [Hevea brasiliensis] |
| 4 |
Hb_002818_030 |
0.0729729022 |
- |
- |
PREDICTED: protein transport protein SEC23 [Jatropha curcas] |
| 5 |
Hb_003494_020 |
0.0756920982 |
- |
- |
ATP-dependent RNA and DNA helicase, putative [Ricinus communis] |
| 6 |
Hb_004916_010 |
0.076224333 |
- |
- |
PREDICTED: uncharacterized protein LOC105631086 [Jatropha curcas] |
| 7 |
Hb_048219_010 |
0.0765533649 |
- |
- |
coronatine-insensitive 1 [Hevea brasiliensis] |
| 8 |
Hb_002234_170 |
0.0824574863 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 5-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_000072_120 |
0.0841450056 |
- |
- |
serine/threonine protein kinase, putative [Ricinus communis] |
| 10 |
Hb_001799_180 |
0.0856532444 |
- |
- |
PREDICTED: serine/threonine-protein kinase D6PK [Jatropha curcas] |
| 11 |
Hb_000331_670 |
0.0860481221 |
- |
- |
PREDICTED: protein tipD [Jatropha curcas] |
| 12 |
Hb_000049_260 |
0.0862158599 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 13 |
Hb_000603_280 |
0.0880975952 |
- |
- |
PREDICTED: uncharacterized protein LOC105638082 [Jatropha curcas] |
| 14 |
Hb_001674_030 |
0.0887116409 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At2g24240 [Jatropha curcas] |
| 15 |
Hb_015807_150 |
0.0894705047 |
- |
- |
actin binding protein, putative [Ricinus communis] |
| 16 |
Hb_015934_040 |
0.0895769839 |
- |
- |
PREDICTED: protein Hook homolog 3-like [Jatropha curcas] |
| 17 |
Hb_007850_090 |
0.0897588575 |
- |
- |
PREDICTED: probable galacturonosyltransferase 11 isoform X1 [Jatropha curcas] |
| 18 |
Hb_012490_080 |
0.0902305965 |
- |
- |
threonine synthase, putative [Ricinus communis] |
| 19 |
Hb_000406_140 |
0.0922315566 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_001856_230 |
0.0930681591 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 8 [Jatropha curcas] |