| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
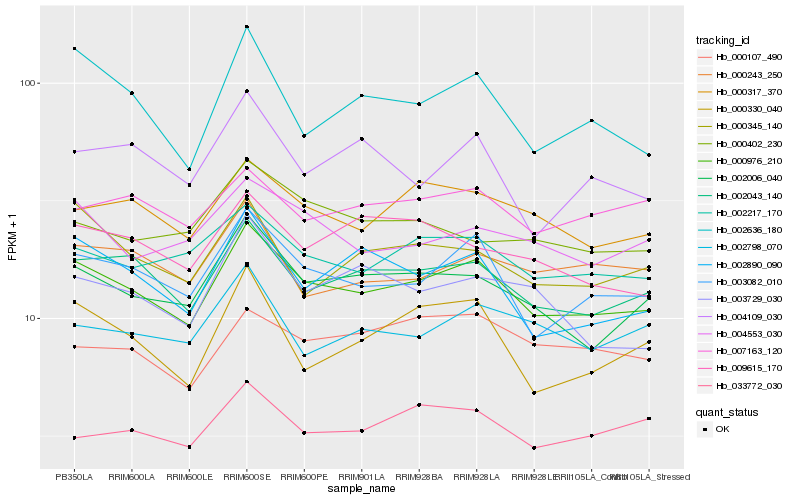
Hb_000976_210 |
0.0 |
- |
- |
PREDICTED: protein PAT1 homolog 1 isoform X1 [Jatropha curcas] |
| 2 |
Hb_002043_140 |
0.0543900413 |
- |
- |
PREDICTED: uncharacterized protein At1g51745-like [Jatropha curcas] |
| 3 |
Hb_003082_010 |
0.0544152455 |
- |
- |
PREDICTED: putative F-box protein At3g10240 [Jatropha curcas] |
| 4 |
Hb_002006_040 |
0.0623174468 |
- |
- |
PREDICTED: translation initiation factor eIF-2B subunit delta isoform X1 [Jatropha curcas] |
| 5 |
Hb_002217_170 |
0.0690812015 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase UPL6-like isoform X1 [Gossypium raimondii] |
| 6 |
Hb_000317_370 |
0.0694284819 |
transcription factor |
TF Family: PHD |
PREDICTED: PHD finger protein At1g33420-like [Jatropha curcas] |
| 7 |
Hb_002890_090 |
0.0730870051 |
- |
- |
Auxin response factor 1 [Glycine soja] |
| 8 |
Hb_004553_030 |
0.077607523 |
- |
- |
PREDICTED: uncharacterized protein LOC105629320 [Jatropha curcas] |
| 9 |
Hb_002636_180 |
0.0800005443 |
- |
- |
inositol or phosphatidylinositol kinase, putative [Ricinus communis] |
| 10 |
Hb_000345_140 |
0.0802435072 |
- |
- |
PREDICTED: autophagy-related protein 18a-like [Jatropha curcas] |
| 11 |
Hb_000330_040 |
0.0808809249 |
- |
- |
PREDICTED: protein RER1B-like [Jatropha curcas] |
| 12 |
Hb_033772_030 |
0.0837641717 |
- |
- |
PREDICTED: CST complex subunit CTC1 [Jatropha curcas] |
| 13 |
Hb_002798_070 |
0.0840816169 |
transcription factor |
TF Family: Jumonji |
PREDICTED: putative lysine-specific demethylase JMJ16 [Jatropha curcas] |
| 14 |
Hb_009615_170 |
0.0842880299 |
- |
- |
PREDICTED: sphingoid long-chain bases kinase 1 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000243_250 |
0.0847961981 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 44 [Jatropha curcas] |
| 16 |
Hb_007163_120 |
0.0856453451 |
- |
- |
PREDICTED: COMPASS-like H3K4 histone methylase component WDR5A [Jatropha curcas] |
| 17 |
Hb_000107_490 |
0.0861504984 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 5 [Jatropha curcas] |
| 18 |
Hb_003729_030 |
0.0865095196 |
- |
- |
PREDICTED: alpha,alpha-trehalose-phosphate synthase [UDP-forming] 6 [Jatropha curcas] |
| 19 |
Hb_000402_230 |
0.0869346242 |
- |
- |
PREDICTED: sorting nexin 2A-like isoform X1 [Jatropha curcas] |
| 20 |
Hb_004109_030 |
0.0870749713 |
- |
- |
conserved hypothetical protein [Ricinus communis] |