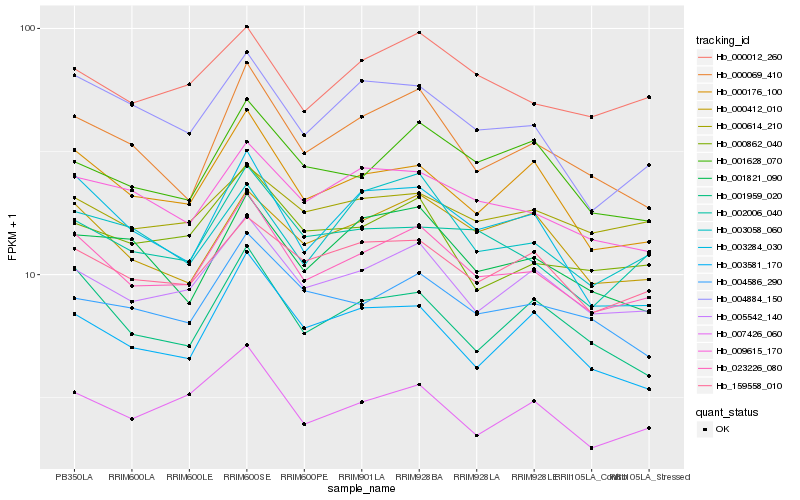
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_023226_080 |
0.0 |
transcription factor |
TF Family: C3H |
PREDICTED: 30-kDa cleavage and polyadenylation specificity factor 30 isoform X1 [Jatropha curcas] |
| 2 |
Hb_005542_140 |
0.0503647925 |
- |
- |
PREDICTED: U-box domain-containing protein 62-like [Jatropha curcas] |
| 3 |
Hb_000176_100 |
0.0595637915 |
- |
- |
PREDICTED: probable splicing factor 3A subunit 1 [Solanum lycopersicum] |
| 4 |
Hb_000069_410 |
0.0649599217 |
- |
- |
PREDICTED: protein SDE2 homolog [Jatropha curcas] |
| 5 |
Hb_159558_010 |
0.0649870761 |
- |
- |
PREDICTED: uncharacterized protein LOC105635846 [Jatropha curcas] |
| 6 |
Hb_000012_260 |
0.0662322096 |
- |
- |
Far upstream element-binding 2 [Gossypium arboreum] |
| 7 |
Hb_009615_170 |
0.0663127424 |
- |
- |
PREDICTED: sphingoid long-chain bases kinase 1 isoform X1 [Jatropha curcas] |
| 8 |
Hb_002006_040 |
0.0667568161 |
- |
- |
PREDICTED: translation initiation factor eIF-2B subunit delta isoform X1 [Jatropha curcas] |
| 9 |
Hb_007426_060 |
0.0690026037 |
desease resistance |
Gene Name: NB-ARC |
unnamed protein product [Coffea canephora] |
| 10 |
Hb_004884_150 |
0.0696722211 |
- |
- |
PREDICTED: ubiquitin-activating enzyme E1 1-like [Jatropha curcas] |
| 11 |
Hb_000412_010 |
0.0711812763 |
- |
- |
hect ubiquitin-protein ligase, putative [Ricinus communis] |
| 12 |
Hb_001959_020 |
0.0718417083 |
- |
- |
PREDICTED: uncharacterized protein LOC105638454 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000862_040 |
0.0720228604 |
- |
- |
ara4-interacting protein, putative [Ricinus communis] |
| 14 |
Hb_004586_290 |
0.0739992806 |
- |
- |
PREDICTED: transport inhibitor response 1-like protein [Jatropha curcas] |
| 15 |
Hb_001628_070 |
0.0744646589 |
- |
- |
hect ubiquitin-protein ligase, putative [Ricinus communis] |
| 16 |
Hb_003581_170 |
0.0760774991 |
- |
- |
PREDICTED: alpha-amylase 3, chloroplastic [Jatropha curcas] |
| 17 |
Hb_003284_030 |
0.076978801 |
transcription factor |
TF Family: Orphans |
PREDICTED: paired amphipathic helix protein Sin3-like 2 isoform X1 [Jatropha curcas] |
| 18 |
Hb_001821_090 |
0.0776906071 |
transcription factor |
TF Family: CAMTA |
PREDICTED: calmodulin-binding transcription activator 2 [Jatropha curcas] |
| 19 |
Hb_000614_210 |
0.0781269112 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor GT-1-like isoform X1 [Jatropha curcas] |
| 20 |
Hb_003058_060 |
0.0788149234 |
- |
- |
PREDICTED: proteasome-associated protein ECM29 homolog [Jatropha curcas] |