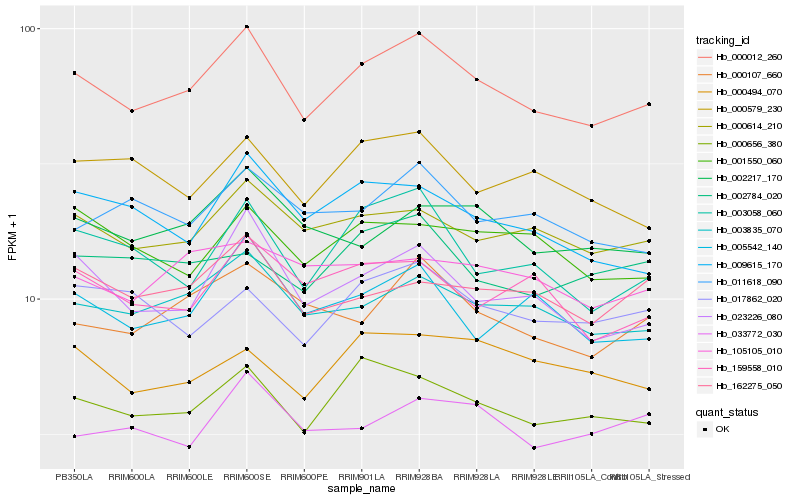
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000012_260 |
0.0 |
- |
- |
Far upstream element-binding 2 [Gossypium arboreum] |
| 2 |
Hb_003835_070 |
0.0595559886 |
- |
- |
PREDICTED: uncharacterized protein LOC105641698 isoform X2 [Jatropha curcas] |
| 3 |
Hb_000656_380 |
0.0605031856 |
- |
- |
PREDICTED: exportin-T [Jatropha curcas] |
| 4 |
Hb_023226_080 |
0.0662322096 |
transcription factor |
TF Family: C3H |
PREDICTED: 30-kDa cleavage and polyadenylation specificity factor 30 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000614_210 |
0.0667787224 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor GT-1-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_003058_060 |
0.0717992495 |
- |
- |
PREDICTED: proteasome-associated protein ECM29 homolog [Jatropha curcas] |
| 7 |
Hb_000107_660 |
0.0731637756 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1 [Jatropha curcas] |
| 8 |
Hb_005542_140 |
0.0754965595 |
- |
- |
PREDICTED: U-box domain-containing protein 62-like [Jatropha curcas] |
| 9 |
Hb_009615_170 |
0.0765003083 |
- |
- |
PREDICTED: sphingoid long-chain bases kinase 1 isoform X1 [Jatropha curcas] |
| 10 |
Hb_002217_170 |
0.0765759439 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase UPL6-like isoform X1 [Gossypium raimondii] |
| 11 |
Hb_159558_010 |
0.0788017172 |
- |
- |
PREDICTED: uncharacterized protein LOC105635846 [Jatropha curcas] |
| 12 |
Hb_162275_050 |
0.0790814352 |
- |
- |
PREDICTED: mRNA-capping enzyme [Jatropha curcas] |
| 13 |
Hb_017862_020 |
0.0793472804 |
- |
- |
PREDICTED: probable inactive serine/threonine-protein kinase lvsG [Jatropha curcas] |
| 14 |
Hb_000579_230 |
0.080082366 |
- |
- |
PREDICTED: bifunctional nuclease 1 [Jatropha curcas] |
| 15 |
Hb_011618_090 |
0.0801240246 |
desease resistance |
Gene Name: ABC_tran |
ABC transporter family protein [Hevea brasiliensis] |
| 16 |
Hb_001550_060 |
0.0807902098 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase BSL3 isoform X4 [Jatropha curcas] |
| 17 |
Hb_000494_070 |
0.0826130261 |
- |
- |
PREDICTED: protein ARABIDILLO 1-like [Jatropha curcas] |
| 18 |
Hb_033772_030 |
0.0835521162 |
- |
- |
PREDICTED: CST complex subunit CTC1 [Jatropha curcas] |
| 19 |
Hb_002784_020 |
0.0838163357 |
- |
- |
PREDICTED: probable protein arginine N-methyltransferase 3 [Jatropha curcas] |
| 20 |
Hb_105105_010 |
0.0839049068 |
- |
- |
PREDICTED: coiled-coil domain-containing protein 132 [Jatropha curcas] |