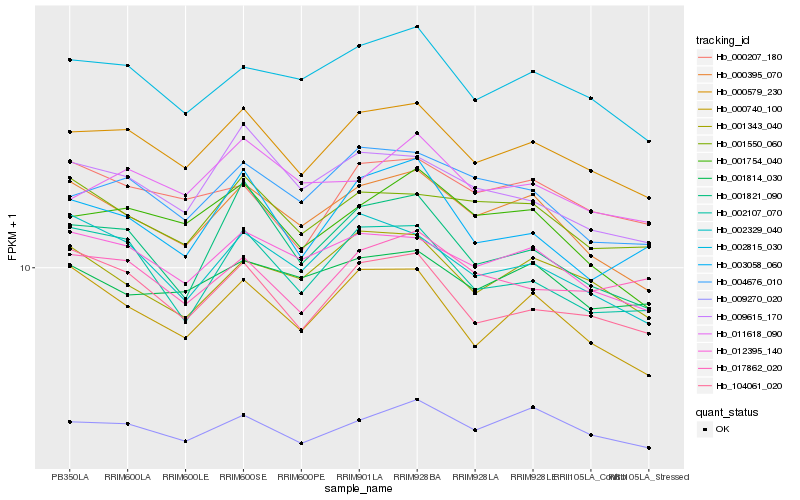
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000579_230 |
0.0 |
- |
- |
PREDICTED: bifunctional nuclease 1 [Jatropha curcas] |
| 2 |
Hb_012395_140 |
0.0492373563 |
- |
- |
PREDICTED: uncharacterized protein LOC105638411 [Jatropha curcas] |
| 3 |
Hb_002815_030 |
0.0492785138 |
- |
- |
hypothetical protein CISIN_1g0095162mg, partial [Citrus sinensis] |
| 4 |
Hb_000395_070 |
0.055257481 |
- |
- |
PREDICTED: serine/threonine-protein kinase BLUS1 isoform X2 [Jatropha curcas] |
| 5 |
Hb_104061_020 |
0.0555110934 |
- |
- |
PREDICTED: UV-stimulated scaffold protein A homolog [Jatropha curcas] |
| 6 |
Hb_009270_020 |
0.0560252845 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: helicase and polymerase-containing protein TEBICHI [Jatropha curcas] |
| 7 |
Hb_004676_010 |
0.0561815255 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g80270, mitochondrial-like [Jatropha curcas] |
| 8 |
Hb_009615_170 |
0.0599056474 |
- |
- |
PREDICTED: sphingoid long-chain bases kinase 1 isoform X1 [Jatropha curcas] |
| 9 |
Hb_001754_040 |
0.0600404225 |
transcription factor |
TF Family: TRAF |
PREDICTED: uncharacterized protein LOC105643434 [Jatropha curcas] |
| 10 |
Hb_001821_090 |
0.0620095356 |
transcription factor |
TF Family: CAMTA |
PREDICTED: calmodulin-binding transcription activator 2 [Jatropha curcas] |
| 11 |
Hb_000207_180 |
0.0637294725 |
- |
- |
PREDICTED: calponin homology domain-containing protein DDB_G0272472 [Jatropha curcas] |
| 12 |
Hb_001550_060 |
0.0637308363 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase BSL3 isoform X4 [Jatropha curcas] |
| 13 |
Hb_000740_100 |
0.0638566622 |
- |
- |
calpain, putative [Ricinus communis] |
| 14 |
Hb_002107_070 |
0.0639263465 |
- |
- |
hypothetical protein RCOM_1598630 [Ricinus communis] |
| 15 |
Hb_002329_040 |
0.0640280678 |
- |
- |
PREDICTED: uncharacterized protein LOC105648065 [Jatropha curcas] |
| 16 |
Hb_011618_090 |
0.0647136964 |
desease resistance |
Gene Name: ABC_tran |
ABC transporter family protein [Hevea brasiliensis] |
| 17 |
Hb_017862_020 |
0.0654612861 |
- |
- |
PREDICTED: probable inactive serine/threonine-protein kinase lvsG [Jatropha curcas] |
| 18 |
Hb_003058_060 |
0.0657249868 |
- |
- |
PREDICTED: proteasome-associated protein ECM29 homolog [Jatropha curcas] |
| 19 |
Hb_001343_040 |
0.0660238932 |
- |
- |
PREDICTED: uncharacterized protein LOC105638555 [Jatropha curcas] |
| 20 |
Hb_001814_030 |
0.066359586 |
- |
- |
PREDICTED: uncharacterized protein LOC105650634 [Jatropha curcas] |