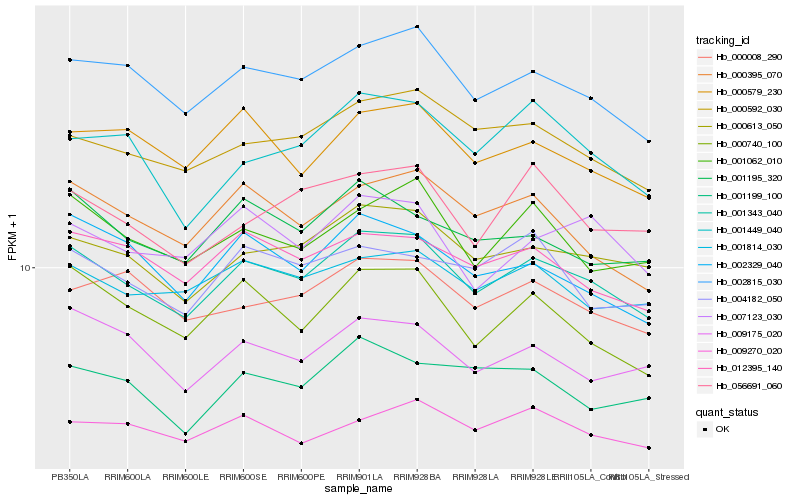
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001343_040 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105638555 [Jatropha curcas] |
| 2 |
Hb_002815_030 |
0.0459409893 |
- |
- |
hypothetical protein CISIN_1g0095162mg, partial [Citrus sinensis] |
| 3 |
Hb_000613_050 |
0.0488120585 |
- |
- |
PREDICTED: uncharacterized protein LOC105641544 isoform X2 [Jatropha curcas] |
| 4 |
Hb_000592_030 |
0.0577399241 |
- |
- |
PREDICTED: uncharacterized protein LOC100853969 isoform X1 [Vitis vinifera] |
| 5 |
Hb_000395_070 |
0.0620426129 |
- |
- |
PREDICTED: serine/threonine-protein kinase BLUS1 isoform X2 [Jatropha curcas] |
| 6 |
Hb_001814_030 |
0.0620680256 |
- |
- |
PREDICTED: uncharacterized protein LOC105650634 [Jatropha curcas] |
| 7 |
Hb_012395_140 |
0.0621314175 |
- |
- |
PREDICTED: uncharacterized protein LOC105638411 [Jatropha curcas] |
| 8 |
Hb_001062_010 |
0.0623601552 |
- |
- |
PREDICTED: CWF19-like protein 2 [Jatropha curcas] |
| 9 |
Hb_000740_100 |
0.0643508405 |
- |
- |
calpain, putative [Ricinus communis] |
| 10 |
Hb_001195_320 |
0.064406705 |
- |
- |
PREDICTED: S-adenosyl-L-methionine-dependent tRNA 4-demethylwyosine synthase [Jatropha curcas] |
| 11 |
Hb_000579_230 |
0.0660238932 |
- |
- |
PREDICTED: bifunctional nuclease 1 [Jatropha curcas] |
| 12 |
Hb_009175_020 |
0.0661604518 |
- |
- |
PREDICTED: uncharacterized protein LOC101303140 [Fragaria vesca subsp. vesca] |
| 13 |
Hb_002329_040 |
0.0663779817 |
- |
- |
PREDICTED: uncharacterized protein LOC105648065 [Jatropha curcas] |
| 14 |
Hb_000008_290 |
0.0672687148 |
- |
- |
PREDICTED: protein SAND [Jatropha curcas] |
| 15 |
Hb_007123_030 |
0.0677695347 |
- |
- |
PREDICTED: uncharacterized protein LOC105634056 isoform X3 [Jatropha curcas] |
| 16 |
Hb_009270_020 |
0.0696214075 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: helicase and polymerase-containing protein TEBICHI [Jatropha curcas] |
| 17 |
Hb_056691_060 |
0.0710839946 |
- |
- |
PREDICTED: probable manganese-transporting ATPase PDR2 isoform X1 [Jatropha curcas] |
| 18 |
Hb_001449_040 |
0.0715718942 |
- |
- |
PREDICTED: SWR1 complex subunit 2 isoform X2 [Jatropha curcas] |
| 19 |
Hb_001199_100 |
0.0719098146 |
- |
- |
PREDICTED: uncharacterized protein LOC105639813 [Jatropha curcas] |
| 20 |
Hb_004182_050 |
0.0719410812 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase BSL3 isoform X2 [Jatropha curcas] |