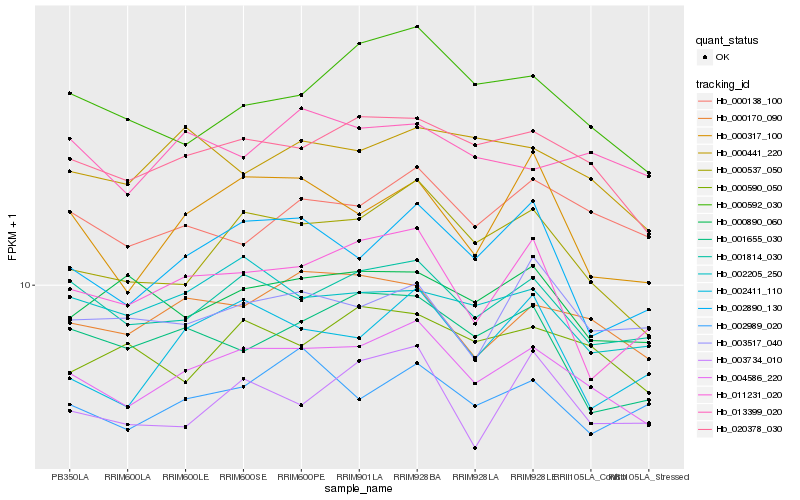
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004586_220 |
0.0 |
- |
- |
PREDICTED: WD and tetratricopeptide repeats protein 1 isoform X3 [Jatropha curcas] |
| 2 |
Hb_020378_030 |
0.0370157894 |
- |
- |
PREDICTED: protein transport protein SEC31 homolog B [Jatropha curcas] |
| 3 |
Hb_001814_030 |
0.0447491036 |
- |
- |
PREDICTED: uncharacterized protein LOC105650634 [Jatropha curcas] |
| 4 |
Hb_000537_050 |
0.0464386167 |
- |
- |
AP-2 complex subunit beta-1, putative [Ricinus communis] |
| 5 |
Hb_000170_090 |
0.0511451225 |
- |
- |
PREDICTED: UDP-sugar pyrophosphorylase [Jatropha curcas] |
| 6 |
Hb_000138_100 |
0.0596242668 |
- |
- |
bifunctional purine biosynthesis protein, putative [Ricinus communis] |
| 7 |
Hb_000441_220 |
0.0604709817 |
- |
- |
PREDICTED: histidinol dehydrogenase, chloroplastic isoform X1 [Jatropha curcas] |
| 8 |
Hb_002205_250 |
0.0612450173 |
- |
- |
PREDICTED: probable inactive serine/threonine-protein kinase scy1 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000592_030 |
0.0613334632 |
- |
- |
PREDICTED: uncharacterized protein LOC100853969 isoform X1 [Vitis vinifera] |
| 10 |
Hb_002890_130 |
0.0614271932 |
- |
- |
tip120, putative [Ricinus communis] |
| 11 |
Hb_002989_020 |
0.0615557565 |
- |
- |
PREDICTED: uncharacterized protein LOC105630013 isoform X2 [Jatropha curcas] |
| 12 |
Hb_000590_050 |
0.0631047727 |
- |
- |
PREDICTED: protein-tyrosine-phosphatase MKP1 [Jatropha curcas] |
| 13 |
Hb_003734_010 |
0.0633960204 |
- |
- |
PREDICTED: double-strand break repair protein MRE11 [Jatropha curcas] |
| 14 |
Hb_000317_100 |
0.0638983176 |
- |
- |
PREDICTED: uncharacterized protein LOC105646995 [Jatropha curcas] |
| 15 |
Hb_013399_020 |
0.0651126697 |
- |
- |
PREDICTED: developmentally-regulated G-protein 2 [Jatropha curcas] |
| 16 |
Hb_001655_030 |
0.0668451247 |
transcription factor |
TF Family: SNF2 |
PREDICTED: SNF2 domain-containing protein CLASSY 3 isoform X2 [Jatropha curcas] |
| 17 |
Hb_000890_060 |
0.066973976 |
- |
- |
PREDICTED: sucrose nonfermenting 4-like protein [Jatropha curcas] |
| 18 |
Hb_003517_040 |
0.0672474978 |
- |
- |
PREDICTED: putative DUF21 domain-containing protein At3g13070, chloroplastic [Jatropha curcas] |
| 19 |
Hb_002411_110 |
0.0672786906 |
- |
- |
PREDICTED: DNA-directed RNA polymerase III subunit RPC2 [Jatropha curcas] |
| 20 |
Hb_011231_020 |
0.0680623932 |
- |
- |
PREDICTED: proline-, glutamic acid- and leucine-rich protein 1 isoform X2 [Jatropha curcas] |