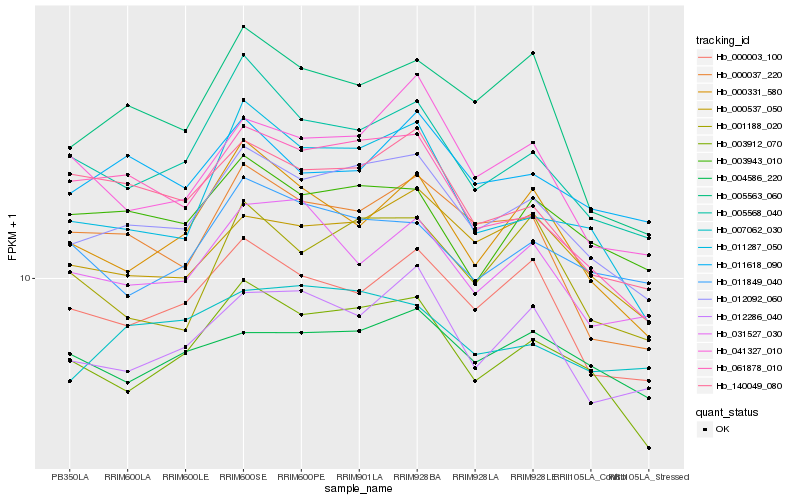
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012092_060 |
0.0 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375 [Jatropha curcas] |
| 2 |
Hb_003912_070 |
0.0492220241 |
- |
- |
PREDICTED: nuclear pore complex protein NUP85 [Jatropha curcas] |
| 3 |
Hb_000537_050 |
0.0598848986 |
- |
- |
AP-2 complex subunit beta-1, putative [Ricinus communis] |
| 4 |
Hb_005568_040 |
0.0612462583 |
- |
- |
PREDICTED: uncharacterized protein LOC105628163 isoform X2 [Jatropha curcas] |
| 5 |
Hb_012286_040 |
0.0629403051 |
- |
- |
PREDICTED: pre-mRNA-splicing factor RSE1 [Jatropha curcas] |
| 6 |
Hb_000003_100 |
0.0636427728 |
transcription factor |
TF Family: SNF2 |
PREDICTED: transcription termination factor 2 isoform X5 [Jatropha curcas] |
| 7 |
Hb_011287_050 |
0.0683237017 |
- |
- |
PREDICTED: uncharacterized protein LOC105644786 [Jatropha curcas] |
| 8 |
Hb_061878_010 |
0.0703178293 |
- |
- |
hypothetical protein CISIN_1g0094072mg, partial [Citrus sinensis] |
| 9 |
Hb_140049_080 |
0.0703667769 |
- |
- |
PREDICTED: probable RNA helicase SDE3 [Jatropha curcas] |
| 10 |
Hb_000037_220 |
0.0708613003 |
- |
- |
PREDICTED: vacuolar protein-sorting-associated protein 11 homolog [Jatropha curcas] |
| 11 |
Hb_000331_580 |
0.0718029906 |
- |
- |
PREDICTED: programmed cell death protein 4-like [Jatropha curcas] |
| 12 |
Hb_003943_010 |
0.0722384897 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_011618_090 |
0.0726082516 |
desease resistance |
Gene Name: ABC_tran |
ABC transporter family protein [Hevea brasiliensis] |
| 14 |
Hb_007062_030 |
0.0729132505 |
- |
- |
hypothetical protein VITISV_039033 [Vitis vinifera] |
| 15 |
Hb_031527_030 |
0.0731143212 |
- |
- |
PREDICTED: ribosomal RNA processing protein 1 homolog [Jatropha curcas] |
| 16 |
Hb_041327_010 |
0.0731350914 |
- |
- |
PREDICTED: la-related protein 1A isoform X1 [Jatropha curcas] |
| 17 |
Hb_005563_060 |
0.0736379512 |
- |
- |
PREDICTED: coatomer subunit alpha-1 [Jatropha curcas] |
| 18 |
Hb_001188_020 |
0.0741613016 |
- |
- |
PREDICTED: E3 SUMO-protein ligase SIZ1 isoform X1 [Jatropha curcas] |
| 19 |
Hb_004586_220 |
0.076298028 |
- |
- |
PREDICTED: WD and tetratricopeptide repeats protein 1 isoform X3 [Jatropha curcas] |
| 20 |
Hb_011849_040 |
0.0769400746 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |